The helical structure of surfactant peptide KL4 when bound to POPC: POPG lipid vesicles
- PMID: 18636713
- PMCID: PMC2629594
- DOI: 10.1021/bi702551c
The helical structure of surfactant peptide KL4 when bound to POPC: POPG lipid vesicles
Abstract
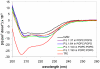
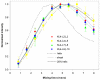
KL 4 is a 21-residue peptide employed as a functional mimic of lung surfactant protein B, which successfully lowers surface tension in the alveoli. A mechanistic understanding of how KL 4 affects lipid properties has proven elusive as the secondary structure of KL 4 in lipid preparations has not been determined at high resolution. The sequence of KL 4 is based on the C-terminus of SP-B, a naturally occurring helical protein that binds to lipid interfaces. The spacing of the lysine residues in KL 4 precludes the formation of a canonical amphipathic alpha-helix; qualitative measurements using Raman, CD, and FTIR spectroscopies have given conflicting results as to the secondary structure of the peptide as well as its orientation in the lipid environment. Here, we present a structural model of KL 4 bound to lipid bilayers based on solid state NMR data. Double-quantum correlation experiments employing (13)C-enriched peptides were used to quantitatively determine the backbone torsion angles in KL 4 at several positions. These measurements, coupled with CD experiments, verify the helical nature of KL 4 when bound to lipids, with (phi, psi) angles that differ substantially from common values for alpha-helices of (-60, -45). The average torsion angles found for KL 4 bound to POPC:POPG lipid vesicles are (-105, -30); this deviation from ideal alpha-helical structure allows KL 4 to form an amphipathic helix at the lipid interface.
Figures




Similar articles
-
Penetration depth of surfactant peptide KL4 into membranes is determined by fatty acid saturation.Biophys J. 2009 May 20;96(10):4085-98. doi: 10.1016/j.bpj.2008.12.3966. Biophys J. 2009. PMID: 19450480 Free PMC article.
-
Partitioning, dynamics, and orientation of lung surfactant peptide KL(4) in phospholipid bilayers.Biochim Biophys Acta. 2010 Feb;1798(2):216-22. doi: 10.1016/j.bbamem.2009.08.020. Epub 2009 Sep 6. Biochim Biophys Acta. 2010. PMID: 19735643 Free PMC article.
-
Interactions of the C-terminus of lung surfactant protein B with lipid bilayers are modulated by acyl chain saturation.Biochim Biophys Acta. 2008 Nov;1778(11):2544-54. doi: 10.1016/j.bbamem.2008.07.013. Epub 2008 Jul 25. Biochim Biophys Acta. 2008. PMID: 18694722 Free PMC article.
-
Solid-state nuclear magnetic resonance relaxation studies of the interaction mechanism of antimicrobial peptides with phospholipid bilayer membranes.Biochemistry. 2005 Aug 2;44(30):10208-17. doi: 10.1021/bi050730p. Biochemistry. 2005. PMID: 16042398
-
Structure and dynamics of phospholipids in membranes elucidated by combined use of NMR and vibrational spectroscopies.Biochim Biophys Acta Biomembr. 2020 Sep 1;1862(9):183352. doi: 10.1016/j.bbamem.2020.183352. Epub 2020 May 11. Biochim Biophys Acta Biomembr. 2020. PMID: 32407775 Review.
Cited by
-
Membrane protein structure determination: back to the membrane.Methods Mol Biol. 2013;1063:145-58. doi: 10.1007/978-1-62703-583-5_8. Methods Mol Biol. 2013. PMID: 23975776 Free PMC article.
-
Penetration depth of surfactant peptide KL4 into membranes is determined by fatty acid saturation.Biophys J. 2009 May 20;96(10):4085-98. doi: 10.1016/j.bpj.2008.12.3966. Biophys J. 2009. PMID: 19450480 Free PMC article.
-
Partitioning, dynamics, and orientation of lung surfactant peptide KL(4) in phospholipid bilayers.Biochim Biophys Acta. 2010 Feb;1798(2):216-22. doi: 10.1016/j.bbamem.2009.08.020. Epub 2009 Sep 6. Biochim Biophys Acta. 2010. PMID: 19735643 Free PMC article.
-
Interactions of the C-terminus of lung surfactant protein B with lipid bilayers are modulated by acyl chain saturation.Biochim Biophys Acta. 2008 Nov;1778(11):2544-54. doi: 10.1016/j.bbamem.2008.07.013. Epub 2008 Jul 25. Biochim Biophys Acta. 2008. PMID: 18694722 Free PMC article.
-
Probing ground and excited states of phospholamban in model and native lipid membranes by magic angle spinning NMR spectroscopy.Biochim Biophys Acta. 2012 Feb;1818(2):146-53. doi: 10.1016/j.bbamem.2011.07.040. Epub 2011 Aug 3. Biochim Biophys Acta. 2012. PMID: 21839724 Free PMC article.
References
-
- Goerke J. Pulmonary surfactant: functions and molecular composition. Biochimica Et Biophysica Acta-Molecular Basis of Disease. 1998;1408:79–89. - PubMed
-
- Piknova B, Schram V, Hall SB. Pulmonary surfactant: phase behavior and function. Curr Opin Struct Biol. 2002;12:487–494. - PubMed
-
- Whitsett JA, Weaver TE. Mechanisms of disease: Hydrophobic surfactant proteins in lung function and disease. New England Journal of Medicine. 2002;347:2141–2148. - PubMed
-
- Wright JR. Immunoregulatory functions of surfactant proteins. Nature Reviews Immunology. 2005;5:58–68. - PubMed
-
- Serrano AG, Perez-Gil J. Protein-lipid interactions and surface activity in the pulmonary surfactant system. Chem Phys Lipids. 2006;141:105–118. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources

