Antagonistic pleiotropy and p53
- PMID: 18639575
- PMCID: PMC2771578
- DOI: 10.1016/j.mad.2008.06.002
Antagonistic pleiotropy and p53
Abstract
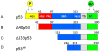
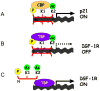
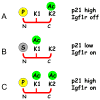
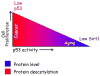
George Williams' antagonistic pleiotropy theory of aging proposes that cellular damage and organismal aging are caused by pleiotrophic genes, or genes with multiple phenotypic effects [Williams, G.C., 1957. Pleiotropy, natural selection, and the evolution of senescence. Evolution 11, 398-411]. According to this theory, genes that exhibit antagonistic pleiotropy increase the odds of successful reproduction early in life, but have deleterious effects later in life. The tumor suppressor p53 confers protection against cancer (and death) by interrupting the abnormal proliferation of cells. When control of proliferation is applied to normal stem cells, however, it can impair tissue homeostasis and accelerate aging. We use data from recently developed models of accelerated aging in mice to determine if the deleterious effects of p53 on aging reflect antagonistic pleiotropy of the p53 gene or are attributable to genes that can modify p53 activity but are evolving independently.
Figures
References
-
- Abramovitch S, Glaser T, Ouchi T, Werner H. BRCA1-Sp1 interactions in transcriptional regulation of the IGF-IR gene. FEBS Lett. 2003;541:149–54. - PubMed
-
- Avantaggiati ML, Ogryzko V, Gardner K, Giordano A, Levine AS, Kelly K. Recruitment of p300/CBP in p53-dependent signal pathways. Cell. 1997;89:1175–84. - PubMed
-
- Baserga RL. Haystacks and needles. Hum Pathol. 2000;31:275–6. - PubMed
-
- Baur JA, Pearson KJ, Price NL, Jamieson HA, Lerin C, Kalra A, Prabhu VV, Allard JS, Lopez-Lluch G, Lewis K, Pistell PJ, Poosala S, Becker KG, Boss O, Gwinn D, Wang M, Ramaswamy S, Fishbein KW, Spencer RG, Lakatta EG, Le Couteur D, Shaw RJ, Navas P, Puigserver P, Ingram DK, de Cabo R, Sinclair DA. Resveratrol improves health and survival of mice on a high-calorie diet. Nature. 2006;444:337–42. - PMC - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
Research Materials
Miscellaneous

