Computationally Optimised DNA Assembly of synthetic genes
- PMID: 18640907
- PMCID: PMC2668710
- DOI: 10.1504/IJBRA.2008.019578
Computationally Optimised DNA Assembly of synthetic genes
Abstract
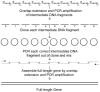
Gene synthesis is hampered by two obstacles: improper assembly of oligonucleotides; oligonucleotide defects incurred during chemical synthesis. To overcome the first problem, we describe the employment of a Computationally Optimised DNA Assembly (CODA) algorithm that uses the degeneracy of the genetic code to design overlapping oligonucleotides with thermodynamic properties for self-assembly into a single, linear, DNA product. To address the second problem, we describe a hierarchical assembly strategy that reduces the incorporation of defective oligonucleotides into full-length gene constructs. The CODA algorithm and these biological methods enable fast, simple and reliable assemblies of sequence-correct full-length genes.
Figures


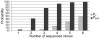
References
-
- Boycheva S, Chkodrov G, Ivanov I. `Codon pairs in the genome of Escherichia coli'. Bioinformatics. 2003;Vol. 19:987–998. - PubMed
-
- Casimiro DR, Wright PE, Dyson HJ. `PCR-based gene synthesis and protein NMR spectroscopy'. Structure. 1997;Vol. 5:1407–1412. - PubMed
-
- Grosjean H, Fiers W. `Preferential codon usage in prokaryotic genes: the optimal codon-anticodon interaction energy and the selective codon usage in efficiently expressed genes'. Gene. 1982;Vol. 18:199–209. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

