Molecular mechanisms underlying the positive stringent response of the Bacillus subtilis ilv-leu operon, involved in the biosynthesis of branched-chain amino acids
- PMID: 18641142
- PMCID: PMC2546780
- DOI: 10.1128/JB.00606-08
Molecular mechanisms underlying the positive stringent response of the Bacillus subtilis ilv-leu operon, involved in the biosynthesis of branched-chain amino acids
Abstract
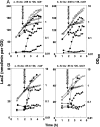
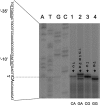
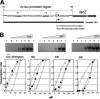
Branched-chain amino acids are the most abundant amino acids in proteins. The Bacillus subtilis ilv-leu operon is involved in the biosynthesis of branched-chain amino acids. This operon exhibits a RelA-dependent positive stringent response to amino acid starvation. We investigated this positive stringent response upon lysine starvation as well as decoyinine treatment. Deletion analysis involving various lacZ fusions revealed two molecular mechanisms underlying the positive stringent response of ilv-leu, i.e., CodY-dependent and -independent mechanisms. The former is most likely triggered by the decrease in the in vivo concentration of GTP upon lysine starvation, GTP being a corepressor of the CodY protein. So, the GTP decrease derepressed ilv-leu expression through detachment of the CodY protein from its cis elements upstream of the ilv-leu promoter. By means of base substitution and in vitro transcription analyses, the latter (CodY-independent) mechanism was found to comprise the modulation of the transcription initiation frequency, which likely depends on fluctuation of the in vivo RNA polymerase substrate concentrations after stringent treatment, and to involve at least the base species of adenine at the 5' end of the ilv-leu transcript. As discussed, this mechanism is presumably distinct from that for B. subtilis rrn operons, which involves changes in the in vivo concentration of the initiating GTP.
Figures





Similar articles
-
Elaborate transcription regulation of the Bacillus subtilis ilv-leu operon involved in the biosynthesis of branched-chain amino acids through global regulators of CcpA, CodY and TnrA.Mol Microbiol. 2005 Jun;56(6):1560-73. doi: 10.1111/j.1365-2958.2005.04635.x. Mol Microbiol. 2005. PMID: 15916606
-
Negative transcriptional regulation of the ilv-leu operon for biosynthesis of branched-chain amino acids through the Bacillus subtilis global regulator TnrA.J Bacteriol. 2004 Dec;186(23):7971-9. doi: 10.1128/JB.186.23.7971-7979.2004. J Bacteriol. 2004. PMID: 15547269 Free PMC article.
-
Activation of the Bacillus subtilis global regulator CodY by direct interaction with branched-chain amino acids.Mol Microbiol. 2004 Jul;53(2):599-611. doi: 10.1111/j.1365-2958.2004.04135.x. Mol Microbiol. 2004. PMID: 15228537
-
Carbon catabolite control of the metabolic network in Bacillus subtilis.Biosci Biotechnol Biochem. 2009 Feb;73(2):245-59. doi: 10.1271/bbb.80479. Epub 2009 Feb 7. Biosci Biotechnol Biochem. 2009. PMID: 19202299 Review.
-
CodY, a master integrator of metabolism and virulence in Gram-positive bacteria.Curr Genet. 2017 Jun;63(3):417-425. doi: 10.1007/s00294-016-0656-5. Epub 2016 Oct 15. Curr Genet. 2017. PMID: 27744611 Review.
Cited by
-
Molecular mechanism and evolution of guanylate kinase regulation by (p)ppGpp.Mol Cell. 2015 Feb 19;57(4):735-749. doi: 10.1016/j.molcel.2014.12.037. Epub 2015 Feb 5. Mol Cell. 2015. PMID: 25661490 Free PMC article.
-
GTP dysregulation in Bacillus subtilis cells lacking (p)ppGpp results in phenotypic amino acid auxotrophy and failure to adapt to nutrient downshift and regulate biosynthesis genes.J Bacteriol. 2014 Jan;196(1):189-201. doi: 10.1128/JB.00918-13. Epub 2013 Oct 25. J Bacteriol. 2014. PMID: 24163341 Free PMC article.
-
Roles of rel(Spn) in stringent response, global regulation and virulence of serotype 2 Streptococcus pneumoniae D39.Mol Microbiol. 2009 May;72(3):590-611. doi: 10.1111/j.1365-2958.2009.06669.x. Mol Microbiol. 2009. PMID: 19426208 Free PMC article.
-
Quantitative phosphoproteome analysis of Bacillus subtilis reveals novel substrates of the kinase PrkC and phosphatase PrpC.Mol Cell Proteomics. 2014 Aug;13(8):1965-78. doi: 10.1074/mcp.M113.035949. Epub 2014 Jan 5. Mol Cell Proteomics. 2014. PMID: 24390483 Free PMC article.
-
Dissecting complex metabolic integration provides direct genetic evidence for CodY activation by guanine nucleotides.J Bacteriol. 2011 Oct;193(20):5637-48. doi: 10.1128/JB.05510-11. Epub 2011 Aug 19. J Bacteriol. 2011. PMID: 21856856 Free PMC article.
References
-
- Barker, M. M., T. Gaal, and R. L. Gourse. 2001. Mechanism of regulation of transcription initiation by ppGpp. II. Models for positive control based on properties of RNAP mutants and competition for RNAP. J. Mol. Biol. 305689-702. - PubMed
-
- Cashel, M., D. R. Gentry, V. J. Hernandez, and D. Vinella. 1996. The stringent response, p. 1458-1496. In F. C. Neidhardt, R. Curtiss III, J. L. Ingraham, E. C. C. Lin, K. B. Low, B. Magasanik, W. S. Reznikoff, M. Riley, M. Schaechter, and E. Umbarger (ed.), Escherichia coli and Salmonella: cellular and molecular biology, 2nd ed. ASM Press, Washington, DC.
-
- de Mendoza, D., G. E. Schujman, and P. S. Aguilar. 2002. Biosynthesis and function of membrane lipids, p. 43-55. In A. L. Sonenshein, J. A. Hoch, and R. Losick (ed.), Bacillus subtilis and its closest relatives: from genes to cells. ASM Press, Washington, DC.
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Research Materials

