Identification of an evolutionarily conserved transcriptional signature of CD8 memory differentiation that is shared by T and B cells
- PMID: 18641323
- PMCID: PMC3771862
- DOI: 10.4049/jimmunol.181.3.1859
Identification of an evolutionarily conserved transcriptional signature of CD8 memory differentiation that is shared by T and B cells
Abstract
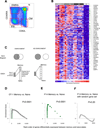
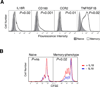
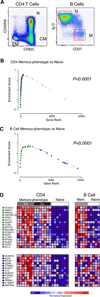
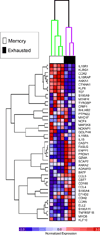
After Ag encounter, naive lymphocytes differentiate into populations of memory cells that share a common set of functions including faster response to Ag re-exposure and the ability to self-renew. However, memory lymphocytes in different lymphocyte lineages are functionally and phenotypically diverse. It is not known whether discrete populations of T and B cells use similar transcriptional programs during differentiation into the memory state. We used cross-species genomic analysis to examine the pattern of genes up-regulated during the differentiation of naive lymphocytes into memory cells in multiple populations of human CD4, CD8, and B cell lymphocytes as well as two mouse models of memory development. We identified and validated a signature of genes that was up-regulated in memory cells compared with naive cells in both human and mouse CD8 memory differentiation, suggesting marked evolutionary conservation of this transcriptional program. Surprisingly, this conserved CD8 differentiation signature was also up-regulated during memory differentiation in CD4 and B cell lineages. To validate the biologic significance of this signature, we showed that alterations in this signature of genes could distinguish between functional and exhausted CD8 T cells from a mouse model of chronic viral infection. Finally, we generated genome-wide microarray data from tetramer-sorted human T cells and showed profound differences in this differentiation signature between T cells specific for HIV and those specific for influenza. Thus, our data suggest that in addition to lineage-specific differentiation programs, T and B lymphocytes use a common transcriptional program during memory development that is disrupted in chronic viral infection.
Figures

References
-
- Masopust D, Kaech SM, Wherry EJ, Ahmed R. The role of programming in memory T-cell development. Curr Opin Immunol. 2004;16:217–225. - PubMed
-
- Bertoletti A, Gehring AJ. The immune response during hepatitis B virus infection. J Gen Virol. 2006;87:1439–1449. - PubMed
-
- McMichael AJ. HIV vaccines. Annu Rev Immunol. 2006;24:227–255. - PubMed
-
- McHeyzer-Williams LJ, McHeyzer-Williams MG. Antigen-specific memory B cell development. Annu Rev Immunol. 2005;23:487–513. - PubMed
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials

