Dynamic transcriptome of Schizosaccharomyces pombe shown by RNA-DNA hybrid mapping
- PMID: 18641648
- PMCID: PMC2538488
- DOI: 10.1038/ng.196
Dynamic transcriptome of Schizosaccharomyces pombe shown by RNA-DNA hybrid mapping
Abstract
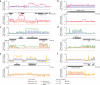
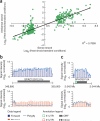
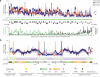
We have determined the high-resolution strand-specific transcriptome of the fission yeast S. pombe under multiple growth conditions using a novel RNA-DNA hybridization mapping (HybMap) technique. HybMap uses an antibody against an RNA-DNA hybrid to detect RNA molecules hybridized to a high-density DNA oligonucleotide tiling microarray. HybMap showed exceptional dynamic range and reproducibility, and allowed us to identify strand-specific coding, noncoding and structural RNAs, as well as previously unknown RNAs conserved in distant yeast species. Notably, we found that virtually the entire euchromatic genome (including intergenics) is transcribed, with heterochromatin dampening intergenic transcription. We identified features including large numbers of condition-specific noncoding RNAs, extensive antisense transcription, new properties of antisense transcripts and induced divergent transcription. Furthermore, our HybMap data informed the efficiency and locations of RNA splicing genome-wide. Finally, we observed strand-specific transcription islands around tRNAs at heterochromatin boundaries inside centromeres. Here, we discuss these new features in terms of organism fitness and transcriptome evolution.
Figures




Comment in
-
Mapping the strand-specific transcriptome of fission yeast.Nat Genet. 2008 Aug;40(8):935-6. doi: 10.1038/ng0808-935. Nat Genet. 2008. PMID: 18665130 No abstract available.
References
-
- Okazaki Y, et al. Analysis of the mouse transcriptome based on functional annotation of 60,770 full-length cDNAs. Nature. 2002;420:563–573. - PubMed
-
- Washietl S, Hofacker IL, Lukasser M, Huttenhofer A, Stadler PF. Mapping of conserved RNA secondary structures predicts thousands of functional noncoding RNAs in the human genome. Nat. Biotechnol. 2005;23:1383–1390. - PubMed
Publication types
MeSH terms
Substances
Associated data
- Actions
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

