Evidence for the bifunctional nature of mitochondrial phosphatidylserine decarboxylase: role in Pdr3-dependent retrograde regulation of PDR5 expression
- PMID: 18644857
- PMCID: PMC2547020
- DOI: 10.1128/MCB.00405-08
Evidence for the bifunctional nature of mitochondrial phosphatidylserine decarboxylase: role in Pdr3-dependent retrograde regulation of PDR5 expression
Abstract
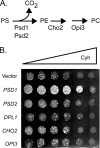
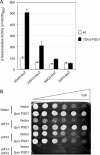
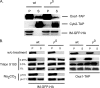
Multidrug resistance in the yeast Saccharomyces cerevisiae is sensitive to the mitochondrial genome status of cells. Cells that lose their organellar genome ([rho(0)] cells) dramatically induce transcription of multiple or pleiotropic drug resistance genes via increased expression of a zinc cluster-containing transcription factor designated Pdr3. A major Pdr3 target gene is the ATP-binding cassette transporter-encoding gene PDR5. Pdr5 has been demonstrated to act as a phospholipid floppase catalyzing the net outward movement of phosphatidylethanolamine (PE). Since the mitochondrially localized Psd1 enzyme provides a major route of PE biosynthesis, we evaluated the potential linkage between Psd1 function and PDR5 regulation. Overproduction of Psd1 in wild-type ([rho(+)]) cells was found to induce PDR5 transcription and drug resistance in a Pdr3-dependent manner. Loss of the PSD1 gene from [rho(0)] cells prevented the normal activation of PDR5 expression. Surprisingly, expression of a catalytically inactive form of Psd1 still supported PDR5 transcriptional activation, suggesting that PE levels were not the signal triggering PDR5 induction. Expression of green fluorescent protein fusions mapped the region required to induce PDR5 expression to the noncatalytic amino-terminal portion of Psd1. Psd1 is a novel bifunctional protein required both for PE biosynthesis and regulation of multidrug resistance.
Figures






Similar articles
-
RPD3 and UME6 are involved in the activation of PDR5 transcription and pleiotropic drug resistance in ρ0 cells of Saccharomyces cerevisiae.BMC Microbiol. 2021 Nov 9;21(1):311. doi: 10.1186/s12866-021-02373-1. BMC Microbiol. 2021. PMID: 34753419 Free PMC article.
-
Expression of an ATP-binding cassette transporter-encoding gene (YOR1) is required for oligomycin resistance in Saccharomyces cerevisiae.Mol Cell Biol. 1995 Dec;15(12):6875-83. doi: 10.1128/MCB.15.12.6875. Mol Cell Biol. 1995. PMID: 8524254 Free PMC article.
-
The ATP-binding cassette multidrug transporter Snq2 of Saccharomyces cerevisiae: a novel target for the transcription factors Pdr1 and Pdr3.Mol Microbiol. 1996 Apr;20(1):109-17. doi: 10.1111/j.1365-2958.1996.tb02493.x. Mol Microbiol. 1996. PMID: 8861209
-
Retrograde regulation of multidrug resistance in Saccharomyces cerevisiae.Gene. 2005 Jul 18;354:15-21. doi: 10.1016/j.gene.2005.03.019. Gene. 2005. PMID: 15896930 Review.
-
Yeast multidrug resistance: the PDR network.J Bioenerg Biomembr. 1995 Feb;27(1):71-6. doi: 10.1007/BF02110333. J Bioenerg Biomembr. 1995. PMID: 7629054 Review.
Cited by
-
Phospholipid biosynthesis disruption renders the yeast cells sensitive to antifungals.Folia Microbiol (Praha). 2020 Feb;65(1):121-131. doi: 10.1007/s12223-019-00713-3. Epub 2019 May 15. Folia Microbiol (Praha). 2020. PMID: 31093957
-
Compartment-specific synthesis of phosphatidylethanolamine is required for normal heavy metal resistance.Mol Biol Cell. 2010 Feb 1;21(3):443-55. doi: 10.1091/mbc.e09-06-0519. Epub 2009 Dec 16. Mol Biol Cell. 2010. PMID: 20016005 Free PMC article.
-
Negative regulation of Candida glabrata Pdr1 by the deubiquitinase subunit Bre5 occurs in a ubiquitin independent manner.Mol Microbiol. 2018 Oct;110(2):309-323. doi: 10.1111/mmi.14109. Epub 2018 Sep 30. Mol Microbiol. 2018. PMID: 30137659 Free PMC article.
-
Identification and Characterization of Key Charged Residues in the Cofilin Protein Involved in Azole Susceptibility, Apoptosis, and Virulence of Aspergillus fumigatus.Antimicrob Agents Chemother. 2018 Apr 26;62(5):e01659-17. doi: 10.1128/AAC.01659-17. Print 2018 May. Antimicrob Agents Chemother. 2018. PMID: 29483117 Free PMC article.
-
Differentially Regulated Transcription Factors and ABC Transporters in a Mitochondrial Dynamics Mutant Can Alter Azole Susceptibility of Aspergillus fumigatus.Front Microbiol. 2020 May 26;11:1017. doi: 10.3389/fmicb.2020.01017. eCollection 2020. Front Microbiol. 2020. PMID: 32528443 Free PMC article.
References
-
- Balzi, E., W. Chen, S. Ulaszewski, E. Capieaux, and A. Goffeau. 1987. The multidrug resistance gene PDR1 from Saccharomyces cerevisiae. J. Biol. Chem. 26216871-16879. - PubMed
-
- Balzi, E., M. Wang, S. Leterme, L. Van Dyck, and A. Goffeau. 1994. PDR5: a novel yeast multidrug resistance transporter controlled by the transcription regulator PDR1. J. Biol. Chem. 2692206-2214. - PubMed
-
- Bevis, B. J., and B. S. Glick. 2002. Rapidly maturing variants of the Discosma red fluorescent protein (DsRed). Nat. Biotechnol. 2083-87. - PubMed
-
- Bissinger, P. H., and K. Kuchler. 1994. Molecular cloning and expression of the S. cerevisiae STS1 gene product. J. Biol. Chem. 2694180-4186. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
