Diadenosine tetraphosphate hydrolase is part of the transcriptional regulation network in immunologically activated mast cells
- PMID: 18644867
- PMCID: PMC2546939
- DOI: 10.1128/MCB.00106-08
Diadenosine tetraphosphate hydrolase is part of the transcriptional regulation network in immunologically activated mast cells
Abstract
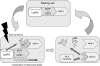
We previously discovered that microphthalmia transcription factor (MITF) and upstream stimulatory factor 2 (USF2) each forms a complex with its inhibitor histidine triad nucleotide-binding 1 (Hint-1) and with lysyl-tRNA synthetase (LysRS). Moreover, we showed that the dinucleotide diadenosine tetraphosphate (Ap(4)A), previously shown to be synthesized by LysRS, binds to Hint-1, and as a result the transcription factors are released from their suppression. Thus, transcriptional activity is regulated by Ap(4)A, suggesting that Ap(4)A is a second messenger in this context. For Ap(4)A to be unambiguously established as a second messenger, several criteria have to be fulfilled, including the presence of a metabolizing enzyme. Since several enzymes are able to hydrolyze Ap(4)A, we provided here evidence that the "Nudix" type 2 gene product, Ap(4)A hydrolase, is responsible for Ap(4)A degradation following the immunological activation of mast cells. The knockdown of Ap(4)A hydrolase modulated Ap(4)A accumulation, resulting in changes in the expression of MITF and USF2 target genes. Moreover, our observations demonstrated that the involvement of Ap(4)A hydrolase in gene regulation is not a phenomenon exclusive to mast cells but can also be found in cardiac cells activated with the beta-agonist isoproterenol. Thus, we have provided concrete evidence establishing Ap(4)A as a second messenger in the regulation of gene expression.
Figures


References
-
- Bessman, M. J., D. N. Frick, and S. F. O'Handley. 1996. The MutT proteins or “Nudix” hydrolases, a family of versatile, widely distributed, “housecleaning” enzymes. J. Biol. Chem. 27125059-25062. - PubMed
-
- Chu, T. Y., and P. Besmer. 1995. Characterization of the promoter of the proto-oncogene c-kit. Proc. Natl. Sci. Counc. Repub. China B 198-18. - PubMed
-
- Fisher, D. I., and A. G. McLennan. 2008. Correlation of intracellular diadenosine triphosphate (Ap3A) with apoptosis in Fhit-positive HEK293 cells. Cancer Lett. 259186-191. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
