An O-phosphotransferase catalyzes phosphorylation of hygromycin A in the antibiotic-producing organism Streptomyces hygroscopicus
- PMID: 18644964
- PMCID: PMC2565881
- DOI: 10.1128/AAC.00157-08
An O-phosphotransferase catalyzes phosphorylation of hygromycin A in the antibiotic-producing organism Streptomyces hygroscopicus
Abstract
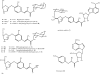
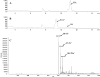
The antibiotic hygromycin A (HA) binds to the 50S ribosomal subunit and inhibits protein synthesis in gram-positive and gram-negative bacteria. The HA biosynthetic gene cluster in Streptomyces hygroscopicus NRRL 2388 contains 29 open reading frames, which have been assigned putative roles in biosynthesis, pathway regulation, and self-resistance. The hyg21 gene encodes an O-phosphotransferase with a proposed role in self-resistance. We observed that insertional inactivation of hyg21 in S. hygroscopicus leads to a greater than 90% decrease in HA production. The wild type and the hyg21 mutant were comparably resistant to HA. Using Escherichia coli as a heterologous host, we expressed and purified Hyg21. Kinetic analyses revealed that the recombinant protein catalyzes phosphorylation of HA (K(m) = 30 +/- 4 microM) at the C-2''' position of the fucofuranose ring in the presence of ATP (K(m) = 200 +/- 20 microM) or GTP (K(m) = 350 +/- 60 microM) with a k(cat) of 2.2 +/- 0.1 min(-1). The phosphorylated HA is inactive against HA-sensitive Delta tolC E. coli and Streptomyces lividans. Hyg21 also phosphorylates methoxyhygromycin A and desmethylenehygromycin A with k(cat) and K(m) values similar to those observed with HA. Phosphorylation of the naturally occurring isomers of 5'''-dihydrohygromycin A and 5'''-dihydromethoxyhygromycin A was about 12 times slower than for the corresponding non-natural isomers. These studies demonstrate that Hyg21 is an O-phosphotransferase with broad substrate specificity, tolerating changes in the aminocyclitol moiety more than in the fucofuranose moiety, and that phosphorylation by Hyg21 is one of several possible mechanisms of self-resistance in S. hygroscopicus NRRL 2388.
Figures




Similar articles
-
Production of hygromycin A analogs in Streptomyces hygroscopicus NRRL 2388 through identification and manipulation of the biosynthetic gene cluster.Chem Biol. 2006 Jul;13(7):753-64. doi: 10.1016/j.chembiol.2006.05.013. Chem Biol. 2006. PMID: 16873023
-
Biosynthesis of the aminocyclitol subunit of hygromycin A in Streptomyces hygroscopicus NRRL 2388.Chem Biol. 2009 Nov 25;16(11):1180-9. doi: 10.1016/j.chembiol.2009.10.013. Chem Biol. 2009. PMID: 19942141
-
The final step of hygromycin A biosynthesis, oxidation of C-5''-dihydrohygromycin A, is linked to a putative proton gradient-dependent efflux.Antimicrob Agents Chemother. 2009 Dec;53(12):5163-72. doi: 10.1128/AAC.01069-09. Epub 2009 Sep 21. Antimicrob Agents Chemother. 2009. PMID: 19770276 Free PMC article.
-
Biochemical basis of resistance to hygromycin B in Streptomyces hygroscopicus--the producing organism.J Gen Microbiol. 1985 Jun;131(6):1289-98. doi: 10.1099/00221287-131-6-1289. J Gen Microbiol. 1985. PMID: 2995542
-
Biosynthetic origin of hygromycin A.Antimicrob Agents Chemother. 2003 Jul;47(7):2065-71. doi: 10.1128/AAC.47.7.2065-2071.2003. Antimicrob Agents Chemother. 2003. PMID: 12821448 Free PMC article.
Cited by
-
Antibiotic resistance and inhibition mechanism of novel aminoglycoside phosphotransferase APH(5) from B. subtilis subsp. subtilis strain RK.Braz J Microbiol. 2019 Oct;50(4):887-898. doi: 10.1007/s42770-019-00132-z. Epub 2019 Aug 10. Braz J Microbiol. 2019. PMID: 31401782 Free PMC article.
-
tRNAs as antibiotic targets.Int J Mol Sci. 2014 Dec 25;16(1):321-49. doi: 10.3390/ijms16010321. Int J Mol Sci. 2014. PMID: 25547494 Free PMC article. Review.
-
Distinct tRNA Accommodation Intermediates Observed on the Ribosome with the Antibiotics Hygromycin A and A201A.Mol Cell. 2015 Jun 4;58(5):832-44. doi: 10.1016/j.molcel.2015.04.014. Epub 2015 May 28. Mol Cell. 2015. PMID: 26028538 Free PMC article.
-
Comparison of Strategies to Overcome Drug Resistance: Learning from Various Kingdoms.Molecules. 2018 Jun 18;23(6):1476. doi: 10.3390/molecules23061476. Molecules. 2018. PMID: 29912169 Free PMC article. Review.
-
Comparison of Antibiotic Resistance Mechanisms in Antibiotic-Producing and Pathogenic Bacteria.Molecules. 2019 Sep 21;24(19):3430. doi: 10.3390/molecules24193430. Molecules. 2019. PMID: 31546630 Free PMC article. Review.
References
-
- Altschul, S. F., W. Gish, W. Miller, E. W. Myers, and D. J. Lipman. 1990. Basic local alignment search tool. J. Mol. Biol. 215:403-410. - PubMed
-
- Barrasa, M. I., J. A. Tercero, and A. Jimenez. 1997. The aminonucleoside antibiotic A201A is inactivated by a phosphotransferase activity from Streptomyces capreolus NRRL 3817, the producing organism: isolation and molecular characterization of the relevant encoding gene and its DNA flanking regions. Eur. J. Biochem. 245:54-63. - PubMed
-
- Butler, A. R., S. A. Flint, and E. Cundliffe. 2001. Feedback control of polyketide metabolism during tylosin production. Microbiology 147:795-801. - PubMed
-
- Cundliffe, E. 1989. How antibiotic-producing organisms avoid suicide. Annu. Rev. Microbiol. 43:207-233. - PubMed
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Medical
Miscellaneous

