Finding of kinase domain mutations in patients with chronic phase chronic myeloid leukemia responding to imatinib may identify those at high risk of disease progression
- PMID: 18645191
- PMCID: PMC10466446
- DOI: 10.1200/JCO.2008.16.9953
Finding of kinase domain mutations in patients with chronic phase chronic myeloid leukemia responding to imatinib may identify those at high risk of disease progression
Abstract
Purpose: Kinase domain (KD) mutations in the BCR-ABL gene are associated with resistance to imatinib in chronic myeloid leukemia (CML) but their incidence and prognostic significance in chronic phase (CP) patients without resistance are unclear.
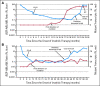
Patients and methods: We analyzed outcome for 319 patients with CML-CP who were treated with imatinib; 171 were in early CP (ECP) and 148 were in late CP (LCP). Patients were screened routinely for mutations using direct sequencing regardless of response status. The 5-year cumulative incidence of mutations was 6.6% for ECP and 17% for LCP patients.
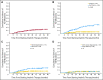
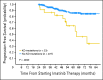
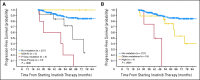
Results: Of the 319 patients, 214 (67%) achieved complete cytogenetic responses (CCyR). The identification of a mutation without other evidence of imatinib resistance was highly predictive for loss of CCyR (RR, 3.8; P = .005) and for progression to advanced phase (RR, 2.3; P = .01), though the intervals from first identification to loss of CCyR and disease progression were relatively long (median, 21 and 16 months, respectively). Mutations in the P-loop (excluding residue 244) were associated with a higher risk of progression than mutations elsewhere.
Conclusion: We conclude that routine mutation screening of patients who appear to be responding to imatinib may identify those at high risk of disease progression.
Conflict of interest statement
Figures
Similar articles
-
Characteristics of BCR-ABL kinase domain point mutations in Chinese imatinib-resistant chronic myeloid leukemia patients.Ann Hematol. 2011 Jan;90(1):47-52. doi: 10.1007/s00277-010-1039-5. Epub 2010 Aug 10. Ann Hematol. 2011. PMID: 20697894
-
Detection of BCR-ABL mutations in patients with CML treated with imatinib is virtually always accompanied by clinical resistance, and mutations in the ATP phosphate-binding loop (P-loop) are associated with a poor prognosis.Blood. 2003 Jul 1;102(1):276-83. doi: 10.1182/blood-2002-09-2896. Epub 2003 Mar 6. Blood. 2003. PMID: 12623848
-
Determining the rise in BCR-ABL RNA that optimally predicts a kinase domain mutation in patients with chronic myeloid leukemia on imatinib.Blood. 2009 Sep 24;114(13):2598-605. doi: 10.1182/blood-2008-08-173674. Epub 2009 Jul 22. Blood. 2009. PMID: 19625707 Free PMC article.
-
Monitoring minimal residual disease in BCR-ABL-positive chronic myeloid leukemia in the imatinib era.Curr Opin Hematol. 2005 Jan;12(1):33-9. doi: 10.1097/01.moh.0000148551.93303.9e. Curr Opin Hematol. 2005. PMID: 15604889 Review.
-
Targeted chronic myeloid leukemia therapy: seeking a cure.J Manag Care Pharm. 2007 Oct;13(8 Suppl A):8-12. doi: 10.18553/jmcp.2007.13.s8-a.8. J Manag Care Pharm. 2007. PMID: 17970609 Free PMC article. Review.
Cited by
-
The BCR-ABLT315I mutation compromises survival in chronic phase chronic myelogenous leukemia patients resistant to tyrosine kinase inhibitors, in a matched pair analysis.Haematologica. 2013 Oct;98(10):1510-6. doi: 10.3324/haematol.2012.080234. Epub 2013 May 28. Haematologica. 2013. PMID: 23716543 Free PMC article.
-
Role of autoimmune hemolytic anemia as an initial indicator for chronic myeloid leukemia: A case report.Medicine (Baltimore). 2020 Feb;99(9):e19256. doi: 10.1097/MD.0000000000019256. Medicine (Baltimore). 2020. PMID: 32118733 Free PMC article.
-
Chronic myeloid leukemia: reminiscences and dreams.Haematologica. 2016 May;101(5):541-58. doi: 10.3324/haematol.2015.139337. Haematologica. 2016. PMID: 27132280 Free PMC article. Review.
-
Exploring treatment decision-making in chronic myeloid leukemia in chronic phase.Front Oncol. 2024 Jul 1;14:1369246. doi: 10.3389/fonc.2024.1369246. eCollection 2024. Front Oncol. 2024. PMID: 39011484 Free PMC article. Review.
-
CML patients in the molecular era - report of five years experience of diagnosis and treatment in a single center.J Med Life. 2013 Sep 15;6(3):319-26. Epub 2013 Sep 25. J Med Life. 2013. PMID: 24146695 Free PMC article.
References
-
- O'Brien SG, Guilhot F, Larson RA, et al: Imatinib compared with interferon and low-dose cytarabine for newly diagnosed chronic-phase chronic myeloid leukemia. N Engl J Med 348::994,2003-1004, - PubMed
-
- Druker B, Guilhot F, O'Brien S, et al: Five-year follow-up of imatinib therapy for newly diagnosed chronic myelogenous leukemia in chronic-phase shows sustained responses and high overall survival. N Engl J Med 355::2408,2006-2417, - PubMed
-
- Kantarjian H, Sawyers C, Hochhaus A, et al: Hematologic and cytogenetic responses to imatinib mesylate in chronic myelogenous leukemia. N Engl J Med 346::645,2002-652, - PubMed
-
- Kantarjian HM, Cortes JE, O'Brien S, et al: Long-term survival benefit and improved complete cytogenetic and molecular response rates with imatinib mesylate in Philadelphia chromosome-positive chronic-phase chronic myeloid leukemia after failure of interferon-alpha. Blood 104::1979,2004-1988, - PubMed
-
- Hochhaus A, Druker B, Sawyers C, et al: Favorable long-term follow-up results over six years for response, survival and safety with imatinib mesylate therapy in chronic phase chronic myeloid leukemia post failure of interferon-alpha treatment. Blood 111::1039,2008-1043, - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Miscellaneous

