Conservation of the S10-spc-alpha locus within otherwise highly plastic genomes provides phylogenetic insight into the genus Leptospira
- PMID: 18648538
- PMCID: PMC2481283
- DOI: 10.1371/journal.pone.0002752
Conservation of the S10-spc-alpha locus within otherwise highly plastic genomes provides phylogenetic insight into the genus Leptospira
Abstract
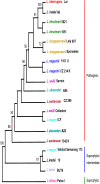
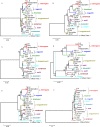
S10-spc-alpha is a 17.5 kb cluster of 32 genes encoding ribosomal proteins. This locus has an unusual composition and organization in Leptospira interrogans. We demonstrate the highly conserved nature of this region among diverse Leptospira and show its utility as a phylogenetically informative region. Comparative analyses were performed by PCR using primer sets covering the whole locus. Correctly sized fragments were obtained by PCR from all L. interrogans strains tested for each primer set indicating that this locus is well conserved in this species. Few differences were detected in amplification profiles between different pathogenic species, indicating that the S10-spc-alpha locus is conserved among pathogenic Leptospira. In contrast, PCR analysis of this locus using DNA from saprophytic Leptospira species and species with an intermediate pathogenic capacity generated varied results. Sequence alignment of the S10-spc-alpha locus from two pathogenic species, L. interrogans and L. borgpetersenii, with the corresponding locus from the saprophyte L. biflexa serovar Patoc showed that genetic organization of this locus is well conserved within Leptospira. Multilocus sequence typing (MLST) of four conserved regions resulted in the construction of well-defined phylogenetic trees that help resolve questions about the interrelationships of pathogenic Leptospira. Based on the results of secY sequence analysis, we found that reliable species identification of pathogenic Leptospira is possible by comparative analysis of a 245 bp region commonly used as a target for diagnostic PCR for leptospirosis. Comparative analysis of Leptospira strains revealed that strain H6 previously classified as L. inadai actually belongs to the pathogenic species L. interrogans and that L. meyeri strain ICF phylogenetically co-localized with the pathogenic clusters. These findings demonstrate that the S10-spc-alpha locus is highly conserved throughout the genus and may be more useful in comparing evolution of the genus than loci studied previously.
Conflict of interest statement
Figures

References
-
- Brenner DJ, Kaufmann AF, Sulzer KR, Steigerwalt AG, Rogers FC, et al. Further determination of DNA relatedness between serogroups and serovars in the family Leptospiraceae with a proposal for Leptospira alexanderi sp. nov. and four new Leptospira genomospecies. Int J Syst Bacteriol 49 Pt. 1999;2:839–858. - PubMed
-
- Ramadass P, Jarvis B, Corner R, Penny D, Marshall R. Genetic characterization of pathogenic Leptospira species by DNA hybridization. Int J Syst Bacteriol. 1992;42:215–219. - PubMed
-
- Yasuda P, Steigerwalt A, Sulzer K, Kaufmann A, Rogers F, et al. Deoxyribonucleic acid relatedness between serogroups and serovars in the family Leptospiraceae with proposals for seven new Leptospira species. Int J Syst Bacteriol. 1987;37:407–415.
-
- Gravekamp C, Van de Kemp H, Franzen M, Carrington D, Schoone GJ, et al. Detection of seven species of pathogenic leptospires by PCR using two sets of primers. J Gen Microbiol. 1993;139:1691–1700. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources

