Diversity and evolution of coral fluorescent proteins
- PMID: 18648549
- PMCID: PMC2481297
- DOI: 10.1371/journal.pone.0002680
Diversity and evolution of coral fluorescent proteins
Abstract
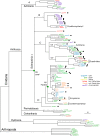
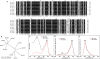
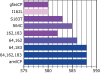
GFP-like fluorescent proteins (FPs) are the key color determinants in reef-building corals (class Anthozoa, order Scleractinia) and are of considerable interest as potential genetically encoded fluorescent labels. Here we report 40 additional members of the GFP family from corals. There are three major paralogous lineages of coral FPs. One of them is retained in all sampled coral families and is responsible for the non-fluorescent purple-blue color, while each of the other two evolved a full complement of typical coral fluorescent colors (cyan, green, and red) and underwent sorting between coral groups. Among the newly cloned proteins are a "chromo-red" color type from Echinopora forskaliana (family Faviidae) and pink chromoprotein from Stylophora pistillata (Pocilloporidae), both evolving independently from the rest of coral chromoproteins. There are several cyan FPs that possess a novel kind of excitation spectrum indicating a neutral chromophore ground state, for which the residue E167 is responsible (numeration according to GFP from A. victoria). The chromoprotein from Acropora millepora is an unusual blue instead of purple, which is due to two mutations: S64C and S183T. We applied a novel probabilistic sampling approach to recreate the common ancestor of all coral FPs as well as the more derived common ancestor of three main fluorescent colors of the Faviina suborder. Both proteins were green such as found elsewhere outside class Anthozoa. Interestingly, a substantial fraction of the all-coral ancestral protein had a chromohore apparently locked in a non-fluorescent neutral state, which may reflect the transitional stage that enabled rapid color diversification early in the history of coral FPs. Our results highlight the extent of convergent or parallel evolution of the color diversity in corals, provide the foundation for experimental studies of evolutionary processes that led to color diversification, and enable a comparative analysis of structural determinants of different colors.
Conflict of interest statement
Figures
References
-
- Lukyanov KA, Chudakov DA, Fradkov AF, Labas YA, Matz MV, et al. Discovery and properties of GFP-like proteins from nonbioluminescent Anthozoa. Methods Biochem Anal. 2006;47:121–138. - PubMed
-
- Chudakov DM, Lukyanov S, Lukyanov KA. Fluorescent proteins as a toolkit for in vivo imaging. Trends in Biotechnology. 2005;23:605–613. - PubMed
-
- Shaner NC, Steinbach PA, Tsien RY. A guide to choosing fluorescent proteins. Nature Methods. 2005;2:905–909. - PubMed
-
- Wachter RM. The family of GFP-like proteins: Structure, function, photophysics and biosensor applications. Photochem Photobiol. 2006;82:339–344. - PubMed
-
- Shagin DA, Barsova EV, Yanushevich YG, Fradkov AF, Lukyanov KA, et al. GFP-like proteins as ubiquitous metazoan superfamily: Evolution of functional features and structural complexity. Mol Biol Evol. 2004;21:841–850. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Miscellaneous

