Metagenomic signatures of the Peru Margin subseafloor biosphere show a genetically distinct environment
- PMID: 18650394
- PMCID: PMC2492506
- DOI: 10.1073/pnas.0709942105
Metagenomic signatures of the Peru Margin subseafloor biosphere show a genetically distinct environment
Abstract
The subseafloor marine biosphere may be one of the largest reservoirs of microbial biomass on Earth and has recently been the subject of debate in terms of the composition of its microbial inhabitants, particularly on sediments from the Peru Margin. A metagenomic analysis was made by using whole-genome amplification and pyrosequencing of sediments from Ocean Drilling Program Site 1229 on the Peru Margin to further explore the microbial diversity and overall community composition within this environment. A total of 61.9 Mb of genetic material was sequenced from sediments at horizons 1, 16, 32, and 50 m below the seafloor. These depths include sediments from both primarily sulfate-reducing methane-generating regions of the sediment column. Many genes of the annotated genes, including those encoding ribosomal proteins, corresponded to those from the Chloroflexi and Euryarchaeota. However, analysis of the 16S small-subunit ribosomal genes suggests that Crenarchaeota are the abundant microbial member. Quantitative PCR confirms that uncultivated Crenarchaeota are indeed a major microbial group in these subsurface samples. These findings show that the marine subsurface is a distinct microbial habitat and is different from environments studied by metagenomics, especially because of the predominance of uncultivated archaeal groups.
Conflict of interest statement
The authors declare no conflict of interest.
Figures
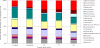

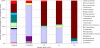

References
-
- D'Hondt S, Rutherford S, Spivack AJ. Metabolic activity of subsurface life in deep-sea sediments. Science. 2002;295:2067–2070. - PubMed
-
- Biddle JF, House CH, Brenchley JE. Enrichment and cultivation of microorganisms from sediment from the slope of the Peru Trench (ODP Site 1230) Proc ODP Sci Results. 2005;201:1–19.
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous

