Global analysis of Arabidopsis gene expression uncovers a complex array of changes impacting pathogen response and cell cycle during geminivirus infection
- PMID: 18650403
- PMCID: PMC2528102
- DOI: 10.1104/pp.108.121038
Global analysis of Arabidopsis gene expression uncovers a complex array of changes impacting pathogen response and cell cycle during geminivirus infection
Abstract
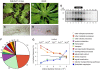
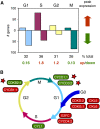
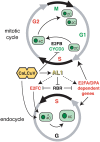
Geminiviruses are small DNA viruses that use plant replication machinery to amplify their genomes. Microarray analysis of the Arabidopsis (Arabidopsis thaliana) transcriptome in response to cabbage leaf curl virus (CaLCuV) infection uncovered 5,365 genes (false discovery rate <0.005) differentially expressed in infected rosette leaves at 12 d postinoculation. Data mining revealed that CaLCuV triggers a pathogen response via the salicylic acid pathway and induces expression of genes involved in programmed cell death, genotoxic stress, and DNA repair. CaLCuV also altered expression of cell cycle-associated genes, preferentially activating genes expressed during S and G2 and inhibiting genes active in G1 and M. A limited set of core cell cycle genes associated with cell cycle reentry, late G1, S, and early G2 had increased RNA levels, while core cell cycle genes linked to early G1 and late G2 had reduced transcripts. Fluorescence-activated cell sorting of nuclei from infected leaves revealed a depletion of the 4C population and an increase in 8C, 16C, and 32C nuclei. Infectivity studies of transgenic Arabidopsis showed that overexpression of CYCD3;1 or E2FB, both of which promote the mitotic cell cycle, strongly impaired CaLCuV infection. In contrast, overexpression of E2FA or E2FC, which can facilitate the endocycle, had no apparent effect. These results showed that geminiviruses and RNA viruses interface with the host pathogen response via a common mechanism, and that geminiviruses modulate plant cell cycle status by differentially impacting the CYCD/retinoblastoma-related protein/E2F regulatory network and facilitating progression into the endocycle.
Figures



References
-
- Accotto GP, Mullineaux PM, Brown SC, Marie D (1993) Digitaria streak geminivirus replicative forms are abundant in S-phase nuclei of infected cells. Virology 195 257–259 - PubMed
-
- Anaya-López JL, Pérez-Mora E, Torres-Pacheco I, Muñoz-Sánchez CI, Guevara-Olvera L, González-Chavira M, Ochoa-Alejo N, Rivera-Bustamante RF, Guevara-González RG (2005) Inducible gene expression by Pepper huasteco virus in Capsicum chinense plants with resistance to geminivirus infections. Can J Plant Pathol 27 276–282
-
- Ascencio-Ibañez JT, Settlage SB (2007) DNA abrasion onto plants is an effective method for geminivirus infection and virus-induced gene silencing. J Virol Methods 142 198–203 - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

