Paclitaxel binding to human and murine MD-2
- PMID: 18650420
- PMCID: PMC2562052
- DOI: 10.1074/jbc.M802826200
Paclitaxel binding to human and murine MD-2
Abstract
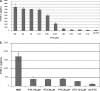
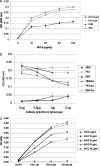
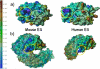
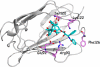
Paclitaxel (PTX) is an important cancer chemotherapeutic agent that binds to beta-tubulin and prevents mitosis through microtubule overstabilization. Recent evidence also implicates PTX in the induction of apoptosis of cancer cells via the TLR4 innate immune pathway. The TLR4 accessory protein, MD-2, is an essential component for the species-specific proinflammatory activity of PTX on murine cells. However, whether PTX binds to human MD-2 and how MD-2 and TLR4 interact with PTX are not well defined. Recombinant human MD-2 (rhMD-2) was produced in a Pichia pastoris expression system, and the interaction between rhMD-2 and PTX was assessed by an enzyme-linked immunosorbent assay to show that PTX binds rhMD-2. Formation of the latter complex was found to be dose-dependent and inhibited by anti-MD-2 antibody but not by an isotype control antibody. As measured by human tumor necrosis factor alpha production, human THP-1 monocytes expressing TLR4 and MD-2 were poorly responsive to the addition of PTX, but murine macrophages expressing TLR4 and MD-2 responded in a dose-dependent manner. Human embryonic kidney (HEK293) cells transfected with both human TLR4 and human MD-2 or human MD-2 and murine TLR4 were also poorly responsive to PTX (10 microm). However, HEK293 cells transfected with murine MD-2 and human TLR4 or murine MD-2 and murine TLR4 were highly responsive to PTX (10 microm), indicating that the murine MD-2/PTX interaction is required for TLR4 activation. To further define the structural differences for MD-2/TLR4 activation, crystal structures of both murine and human MD-2 were subjected to PTX docking by computational methods. These models indicate that PTX binds in the pocket of both human and mouse MD-2 structures. The species-specific difference between human and murine MD-2 activation of TLR4 by PTX can be explained by alterations of surface charge distribution (i.e. electrostatic potential), binding pocket size, and the locus of PTX binding within the MD-2 pocket, which results in reorganization of the 123-130 amino acid loop. In particular, Phe(126) appears to operate as a bridge for TLR4.MD-2 dimerization in the mouse but not the human protein.
Figures



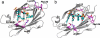

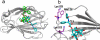


References
-
- Wani, M. C., Taylor, H. L., Wall, M. E., Coggon, P., and McPhail, A. T. (1971) J. Am. Chem. Soc. 93 2325-2327 - PubMed
-
- Schiff, P. B., Fant, J., and Horwitz, S. B. (1979) Nature 277 665-667 - PubMed
-
- Horwitz, S. B., Lothstein, L., Manfredi, J. J., Mellado, W., Parness, J., Roy, S. N., Schiff, P. B., Sorbara, L., and Zeheb, R. (1986) Ann. N. Y. Acad. Sci. 466 733-744 - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Research Materials

