Spatiotemporal regulation of ERK2 by dual specificity phosphatases
- PMID: 18650424
- PMCID: PMC2546534
- DOI: 10.1074/jbc.M801500200
Spatiotemporal regulation of ERK2 by dual specificity phosphatases
Abstract
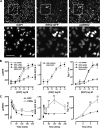
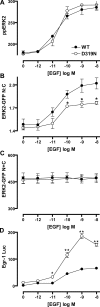
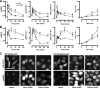
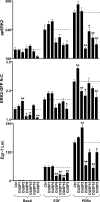
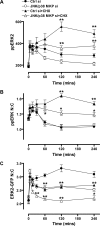
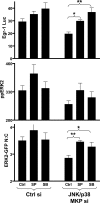
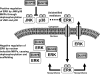
Although many stimuli activate extracellular signal-regulated kinases 1 and 2 (ERK1/2), the kinetics and compartmentalization of ERK1/2 signals are stimulus-dependent and dictate physiological consequences. ERKs can be inactivated by dual specificity phosphatases (DUSPs), notably the MAPK phosphatases (MKPs) and atypical DUSPs, that can both dephosphorylate and scaffold ERK1/2. Using a cell imaging model (based on knockdown of endogenous ERKs and add-back of wild-type or mutated ERK2-GFP reporters), we explored possible effects of DUSPs on responses to transient or sustained ERK2 activators (epidermal growth factor and phorbol 12,13-dibutyrate, respectively). For both stimuli, a D319N mutation (which impairs DUSP binding) increased ERK2 activity and reduced nuclear accumulation. These stimuli also increased mRNA levels for eight DUSPs. In a short inhibitory RNA screen, 12 of 16 DUSPs influenced ERK2 responses. These effects were evident among nuclear inducible MKP, cytoplasmic ERK MKP, JNK/p38 MKP, and atypical DUSP subtypes and, with the exception of the nuclear inducible MKPs, were paralleled by corresponding changes in Egr-1 luciferase activation. Simultaneous removal of all JNK/p38 MKPs or nuclear inducible MKPs revealed them as positive and negative regulators of ERK2 signaling, respectively. The effects of JNK/p38 MKP short inhibitory RNAs were not dependent on protein neosynthesis but were reversed in the presence of JNK and p38 kinase inhibitors, indicating DUSP-mediated cross-talk between MAPK pathways. Overall, our data reveal that a large number of DUSPs influence ERK2 signaling. Together with the known tissue-specific expression of DUSPs and the importance of ERK1/2 in cell regulation, our data support the potential value of DUSPs as targets for drug therapy.
Figures


Similar articles
-
Gonadotropin-releasing hormone and protein kinase C signaling to ERK: spatiotemporal regulation of ERK by docking domains and dual-specificity phosphatases.Mol Endocrinol. 2009 Apr;23(4):510-9. doi: 10.1210/me.2008-0333. Epub 2009 Jan 29. Mol Endocrinol. 2009. PMID: 19179479 Free PMC article.
-
Dual specificity phosphatases 10 and 16 are positive regulators of EGF-stimulated ERK activity: indirect regulation of ERK signals by JNK/p38 selective MAPK phosphatases.Cell Signal. 2012 May;24(5):1002-11. doi: 10.1016/j.cellsig.2011.12.021. Epub 2012 Jan 3. Cell Signal. 2012. PMID: 22245064 Free PMC article.
-
Epidermal growth factor receptor and protein kinase C signaling to ERK2: spatiotemporal regulation of ERK2 by dual specificity phosphatases.J Biol Chem. 2008 Mar 7;283(10):6241-52. doi: 10.1074/jbc.M706624200. Epub 2008 Jan 3. J Biol Chem. 2008. PMID: 18178562 Free PMC article.
-
Dual-specificity phosphatases: critical regulators with diverse cellular targets.Biochem J. 2009 Mar 15;418(3):475-89. doi: 10.1042/bj20082234. Biochem J. 2009. PMID: 19228121 Review.
-
Dual specific phosphatases (DUSPs) in cardiac hypertrophy and failure.Cell Signal. 2021 Aug;84:110033. doi: 10.1016/j.cellsig.2021.110033. Epub 2021 Apr 29. Cell Signal. 2021. PMID: 33933582 Review.
Cited by
-
Pulsatile and sustained gonadotropin-releasing hormone (GnRH) receptor signaling: does the Ca2+/NFAT signaling pathway decode GnRH pulse frequency?J Biol Chem. 2009 Dec 18;284(51):35746-57. doi: 10.1074/jbc.M109.063917. J Biol Chem. 2009. PMID: 19858197 Free PMC article.
-
Induction of Stress Signaling In Vitro and Suppression of Gonadotropin Secretion by Free Fatty Acids in Female Mouse Gonadotropes.Endocrinology. 2018 Feb 1;159(2):1074-1087. doi: 10.1210/en.2017-00638. Endocrinology. 2018. PMID: 29315384 Free PMC article.
-
Mitogen-activated protein kinase phosphatase 2 regulates the inflammatory response in sepsis.Infect Immun. 2010 Jun;78(6):2868-76. doi: 10.1128/IAI.00018-10. Epub 2010 Mar 29. Infect Immun. 2010. PMID: 20351138 Free PMC article.
-
The biology of gonadotroph regulation.Curr Opin Endocrinol Diabetes Obes. 2009 Aug;16(4):321-7. doi: 10.1097/MED.0b013e32832d88fb. Curr Opin Endocrinol Diabetes Obes. 2009. PMID: 19491666 Free PMC article. Review.
-
Long-term dynamics of multisite phosphorylation.Mol Biol Cell. 2016 Jul 15;27(14):2331-40. doi: 10.1091/mbc.E16-03-0137. Epub 2016 May 25. Mol Biol Cell. 2016. PMID: 27226482 Free PMC article.
References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Research Materials
Miscellaneous

