Detecting bacteria and determining their susceptibility to antibiotics by stochastic confinement in nanoliter droplets using plug-based microfluidics
- PMID: 18651067
- PMCID: PMC2612531
- DOI: 10.1039/b804911d
Detecting bacteria and determining their susceptibility to antibiotics by stochastic confinement in nanoliter droplets using plug-based microfluidics
Abstract
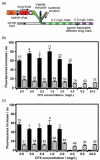
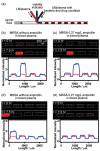
This article describes plug-based microfluidic technology that enables rapid detection and drug susceptibility screening of bacteria in samples, including complex biological matrices, without pre-incubation. Unlike conventional bacterial culture and detection methods, which rely on incubation of a sample to increase the concentration of bacteria to detectable levels, this method confines individual bacteria into droplets nanoliters in volume. When single cells are confined into plugs of small volume such that the loading is less than one bacterium per plug, the detection time is proportional to plug volume. Confinement increases cell density and allows released molecules to accumulate around the cell, eliminating the pre-incubation step and reducing the time required to detect the bacteria. We refer to this approach as 'stochastic confinement'. Using the microfluidic hybrid method, this technology was used to determine the antibiogram - or chart of antibiotic sensitivity - of methicillin-resistant Staphylococcus aureus (MRSA) to many antibiotics in a single experiment and to measure the minimal inhibitory concentration (MIC) of the drug cefoxitin (CFX) against this strain. In addition, this technology was used to distinguish between sensitive and resistant strains of S. aureus in samples of human blood plasma. High-throughput microfluidic techniques combined with single-cell measurements also enable multiple tests to be performed simultaneously on a single sample containing bacteria. This technology may provide a method of rapid and effective patient-specific treatment of bacterial infections and could be extended to a variety of applications that require multiple functional tests of bacterial samples on reduced timescales.
Figures


References
-
- Martin GS, Mannino DM, Eaton S, Moss M. New Engl. J. Med. 2003;348:1546–1554. - PubMed
-
- Farr BM. Curr. Opin. Infect. Dis. 2004;17:317–322. - PubMed
-
- Moran GJ, Krishnadasan A, Gorwitz RJ, Fosheim GE, McDougal LK, Carey RB, Talan DA. New Engl. J. Med. 2006;355:666–674. - PubMed
-
- Carrigan SD, Scott G, Tabrizian M. Clin. Chem. (Washington, D. C.) 2004;50:1301–1314. - PubMed
-
- Nguyen HB, Rivers EP, Abrahamian FM, Moran GJ, Abraham E, Trzeciak S, Huang DT, Osborn T, Stevens D, Talan DA. Ann. Emerg. Med. 2006;48:28–54. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical

