Isolation and expression of Pax6 and atonal homologues in the American horseshoe crab, Limulus polyphemus
- PMID: 18651657
- PMCID: PMC2577597
- DOI: 10.1002/dvdy.21634
Isolation and expression of Pax6 and atonal homologues in the American horseshoe crab, Limulus polyphemus
Abstract
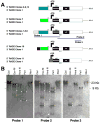
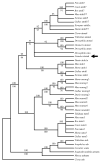
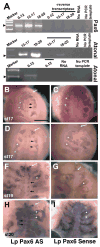
Pax6 regulates eye development in many animals. In addition, Pax6 activates atonal transcription factors in both invertebrate and vertebrate eyes. Here, we investigate the roles of Pax6 and atonal during embryonic development of Limulus polyphemus rudimentary lateral, medial and ventral eyes, and the initiation of lateral ommatidial eye and medial ocelli formation. Limulus eye development is of particular interest because these animals hold a unique position in arthropod phylogeny and possess multiple eye types. Furthermore, the molecular underpinnings of eye development have yet to be investigated in chelicerates. We characterized a Limulus Pax6 gene, with multiple splice products and predicted protein isoforms, and one atonal homologue. Unexpectedly, neither gene is expressed in the developing eye types examined, although both genes are present in the lateral sense organ, a structure of unknown function.
Copyright (c) 2008 Wiley-Liss, Inc.
Figures




Similar articles
-
The beta subunit of voltage-gated Ca2+ channels interacts with and regulates the activity of a novel isoform of Pax6.J Biol Chem. 2010 Jan 22;285(4):2527-36. doi: 10.1074/jbc.M109.022236. Epub 2009 Nov 16. J Biol Chem. 2010. PMID: 19917615 Free PMC article.
-
Temporal and spatial distribution of PAX6 gene expression in the developing human eye.Dokl Biol Sci. 2009 May-Jun;426:264-6. doi: 10.1134/s0012496609030211. Dokl Biol Sci. 2009. PMID: 19650334 No abstract available.
-
Duplicated Pax6 genes in Glomeris marginata (Myriapoda: Diplopoda), an arthropod with simple lateral eyes.Zoology (Jena). 2005;108(1):47-53. doi: 10.1016/j.zool.2004.11.003. Epub 2005 Jan 20. Zoology (Jena). 2005. PMID: 16351954
-
Pax6 3' deletion results in aniridia, autism and mental retardation.Hum Genet. 2008 May;123(4):371-8. doi: 10.1007/s00439-008-0484-x. Epub 2008 Mar 6. Hum Genet. 2008. PMID: 18322702 Free PMC article. Review.
-
Role of a transcription factor Pax6 in the developing vertebrate olfactory system.Dev Growth Differ. 2007 Dec;49(9):683-90. doi: 10.1111/j.1440-169X.2007.00965.x. Epub 2007 Oct 1. Dev Growth Differ. 2007. PMID: 17908181 Review.
Cited by
-
Eye evolution: common use and independent recruitment of genetic components.Philos Trans R Soc Lond B Biol Sci. 2009 Oct 12;364(1531):2819-32. doi: 10.1098/rstb.2009.0079. Philos Trans R Soc Lond B Biol Sci. 2009. PMID: 19720647 Free PMC article. Review.
-
Molecular characterization and embryonic origin of the eyes in the common house spider Parasteatoda tepidariorum.Evodevo. 2015 Apr 28;6:15. doi: 10.1186/s13227-015-0011-9. eCollection 2015. Evodevo. 2015. PMID: 26034574 Free PMC article.
-
A critical analysis of Atoh7 (Math5) mRNA splicing in the developing mouse retina.PLoS One. 2010 Aug 24;5(8):e12315. doi: 10.1371/journal.pone.0012315. PLoS One. 2010. PMID: 20808762 Free PMC article.
-
The expression of Pax6 and retinal determination genes in the eyeless arachnid A. longisetosus reveals vestigial eye primordia.Evodevo. 2025 Jul 9;16(1):12. doi: 10.1186/s13227-025-00245-7. Evodevo. 2025. PMID: 40635118 Free PMC article.
-
Ancestral and novel roles of Pax family genes in mollusks.BMC Evol Biol. 2017 Mar 16;17(1):81. doi: 10.1186/s12862-017-0919-x. BMC Evol Biol. 2017. PMID: 28302062 Free PMC article.
References
-
- Abzhanov A, Popadic A, Kaufman TC. Chelicerate Hox genes and the homology of arthropod segments. Evol Dev. 1999;1:77–89. - PubMed
-
- Arendt D. Evolution of eyes and photoreceptor cell types. Int J Dev Biol. 2003;47:563–571. - PubMed
-
- Arendt D, Tessmar K, de Campos-Baptista MI, Dorresteijn A, Wittbrodt J. Development of pigment-cup eyes in the polychaete Platynereis dumerilii and evolutionary conservation of larval eyes in Bilateria. Development. 2002;129:1143–1154. - PubMed
-
- Avise JC, Nelson WS, Sugita H. A special history of “living fossils”: molecular evolutionary patterns in horseshoe crabs. Evolution. 1994;48:1986–2001. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources

