Intrinsic disorder in nuclear hormone receptors
- PMID: 18651760
- PMCID: PMC2700763
- DOI: 10.1021/pr8003024
Intrinsic disorder in nuclear hormone receptors
Abstract
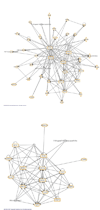
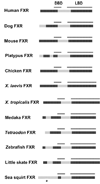
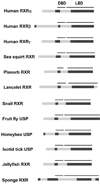
Many proteins possess intrinsic disorder (ID) and lack a rigid three-dimensional structure in at least part of their sequence. ID has been hypothesized to influence protein-protein and protein-ligand interactions. We calculated ID for nearly 400 vertebrate and invertebrate members of the biomedically important nuclear hormone receptor (NHR) superfamily, including all 48 known human NHRs. The predictions correctly identified regions in 20 of the 23 NHRs suggested as disordered based on published X-ray and NMR structures. Of the four major NHR domains (N-terminal domain, DNA-binding domain, D-domain, and ligand-binding domain), we found ID to be highest in the D-domain, a region of NHRs critical in DNA recognition and heterodimerization, coactivator/corepressor interactions and protein-protein interactions. ID in the D-domain and LBD was significantly higher in "hub" human NHRs that have 10 or more downstream proteins in their interaction networks compared to "non-hub" NHRs that interact with fewer than 10 downstream proteins. ID in the D-domain and LBD was also higher in classic, ligand-activated NHRs than in orphan, ligand-independent NHRs in human. The correlation between ID in human and mouse NHRs was high. Less correlation was found for ID between mammalian and non-mammalian vertebrate NHRs. For some invertebrate species, particularly sea squirts ( Ciona), marked differences were observed in ID between invertebrate NHRs and their vertebrate orthologs. Our results indicate that variability of ID within NHRs, particularly in the D-domain and LBD, is likely an important evolutionary force in shaping protein-protein interactions and NHR function. This information enables further understanding of these therapeutic targets.
Figures







References
-
- Dunker AK, Lawson JD, Brown CJ, Williams RM, Romero P, Oh JS, Oldfield CJ, Campen AM, Ratliff CM, Hipps KW, Ausio J, Nissen MS, Reeves R, Kang C, Kissinger CR, Bailey RW, Griswold MD, Chiu W, Garner EC, Obradovic Z. Intrinsically disordered protein. J Mol Graph Model. 2001;19(1):26–59. - PubMed
-
- Dunker AK, Brown CJ, Lawson JD, Iakoucheva LM, Obradovic Z. Intrinsic disorder and protein function. Biochemistry. 2002;41(21):6573–6582. - PubMed
-
- Iakoucheva LM, Brown CJ, Lawson JD, Obradovic Z, Dunker AK. Intrinsic disorder in cell-signaling and cancer-associated proteins. J Mol Biol. 2002;323(3):573–584. - PubMed
-
- Dunker AK, Cortese MS, Romero P, Iakoucheva LM, Uversky VN. Flexible nets. The roles of intrinsic disorder in protein interaction networks. Febs J. 2005;272(20):5129–5148. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources

