Probability that a two-stage genome-wide association study will detect a disease-associated snp and implications for multistage designs
- PMID: 18652601
- PMCID: PMC2574571
- DOI: 10.1111/j.1469-1809.2008.00467.x
Probability that a two-stage genome-wide association study will detect a disease-associated snp and implications for multistage designs
Abstract
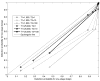
Large two-stage genome-wide association studies (GWASs) have been shown to reduce required genotyping with little loss of power, compared to a one-stage design, provided a substantial fraction of cases and controls, pi(sample), is included in stage 1. However, a number of recent GWASs have used pi(sample) < 0.2. Moreover, standard power calculations are not applicable because SNPs are selected in stage 1 by ranking their p-values, rather than comparing each SNP's statistic to a fixed critical value. We define the detection probability (DP) of a two-stage design as the probability that a given disease-associated SNP will have a p-value among the lowest ranks of p-values at stage 1, and, among those SNPs selected at stage 1, at stage 2. For 8000 cases and 8000 controls available for study and for odds ratios per allele in the range 1.1-1.3, we show that DP is substantially reduced for designs with pi(sample)<or= 0.25, and that DP cannot be appreciably increased by analyzing the stage 1 and stage 2 data jointly. These results suggest that multistage designs with small first stages (e.g. pi(sample)<or= 0.25) should be avoided, and that additional genotyping in earlier studies with small first stages will yield previously unselected disease-associated SNPs.
Figures



References
-
- Aptec Systems. The Gauss System, Version 6. Maple Valley, WA: 2005.
-
- Armitage P. Tests for linear trends in proportions and frequencies. Biometrics. 1955;11:375–386.
-
- Broderick P, Carvajal-Carmona L, Pittman AM, Webb E, Howarth K, Rowan A, Lubbe S, Spain S, Sullivan K, Fielding S, Jaeger E, Vijayakrishnan J, Kemp Z, Gorman M, Chandler I, Papaemmanuil E, Penegar S, Wood W, Sellick G, Qureshi M, Teixeira A, Domingo E, Barclay E, Martin L, Sieber O, Kerr D, Gray R, Peto J, Cazier JB, Tomlinson I, Houlston RS. A genome-wide association study shows that common alleles of SMAD7 influence colorectal cancer risk. Nat Genet. 2007;39:1315–1317. - PubMed
-
- Buch S, Schafmayer C, Volzke H, Becker C, Franke A, Von Eller-Eberstein H, Kluck C, Bassmann I, Brosch M, Lammert F, Miquel JF, Nervi F, Wittig M, Rosskopf D, Timm B, Holl C, Seeger M, Elsharawy A, Lu T, Egberts J, Fandrich F, Folsch UR, Krawczak M, Schreiber S, Nurnberg P, Tepel J, Hampe J. A genome-wide association scan identifies the hepatic cholesterol transporter ABCG8 as a susceptibility factor for human gallstone disease. Nat Genet. 2007;39:995–999. - PubMed
-
- Devlin B, Roeder K. Genomic control for association studies. Biometrics. 1999;55:997–1004. - PubMed
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Research Materials
Miscellaneous

