30 nm chromatin fibre decompaction requires both H4-K16 acetylation and linker histone eviction
- PMID: 18653199
- PMCID: PMC3870898
- DOI: 10.1016/j.jmb.2008.04.050
30 nm chromatin fibre decompaction requires both H4-K16 acetylation and linker histone eviction
Abstract
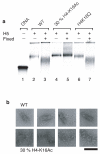
The mechanism by which chromatin is decondensed to permit access to DNA is largely unknown. Here, using a model nucleosome array reconstituted from recombinant histone octamers, we have defined the relative contribution of the individual histone octamer N-terminal tails as well as the effect of a targeted histone tail acetylation on the compaction state of the 30 nm chromatin fiber. This study goes beyond previous studies as it is based on a nucleosome array that is very long (61 nucleosomes) and contains a stoichiometric concentration of bound linker histone, which is essential for the formation of the 30 nm chromatin fiber. We find that compaction is regulated in two steps: Introduction of H4 acetylated to 30% on K16 inhibits compaction to a greater degree than deletion of the H4 N-terminal tail. Further decompaction is achieved by removal of the linker histone.
Figures



References
-
- Kornberg RD. Structure of chromatin. Annu Rev Biochem. 1977;46:931–54. - PubMed
-
- Richmond TJ, Finch JT, Rushton B, Rhodes D, Klug A. Structure of the nucleosome core particle at 7 A resolution. Nature. 1984;311:532–7. - PubMed
-
- Luger K, Mader AW, Richmond RK, Sargent DF, Richmond TJ. Crystal structure of the nucleosome core particle at 2.8 A resolution. Nature. 1997;389:251–60. - PubMed
-
- Allan J, Hartman PG, Crane-Robinson C, Aviles FX. The structure of histone H1 and its location in chromatin. Nature. 1980;288:675–9. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials

