Transcriptional control of spliced and unspliced human T-cell leukemia virus type 1 bZIP factor (HBZ) gene
- PMID: 18653454
- PMCID: PMC2546946
- DOI: 10.1128/JVI.00242-08
Transcriptional control of spliced and unspliced human T-cell leukemia virus type 1 bZIP factor (HBZ) gene
Abstract
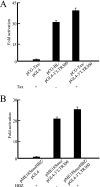
The human T-cell leukemia virus type 1 (HTLV-1) basic leucine zipper factor (HBZ) gene is encoded by the minus strand of the HTLV-1 provirus and transcribed from the 3' long terminal repeat (LTR). HBZ gene expression not only inhibits the Tax-mediated activation of viral gene transcription through the 5' LTR but also promotes the proliferation of infected cells. However, the HBZ promoter region and the transcriptional regulation of the gene have not been studied. In this study, we characterize the promoters of the spliced version of the HBZ gene (sHBZ) and the unspliced version of the HBZ gene (usHBZ) by luciferase assay. Both promoters were TATA-less and contained initiators and downstream promoter elements. Detailed studies of the promoter for the sHBZ gene showed that Sp1 sites were critical for its activity. The activities of the sHBZ and usHBZ gene promoters were upregulated by Tax through Tax-responsible elements in the 3' LTR. We compared the functions of the proteins derived from the sHBZ and usHBZ transcripts. sHBZ showed a stronger suppression of Tax-mediated transcriptional activation through the 5' LTR than did usHBZ; the level of suppression correlated with the level of protein produced. The expression of sHBZ had a growth-promoting function in a T-cell line, while usHBZ expression did not. This study demonstrates that Sp1 is critical for sHBZ transcription, which accounts for the constitutive expression of the sHBZ gene. Functional differences between sHBZ and usHBZ suggest that the sHBZ gene plays a significant role in the proliferation of infected cells.
Figures





References
-
- Basbous, J., C. Arpin, G. Gaudray, M. Piechaczyk, C. Devaux, and J. M. Mesnard. 2003. The HBZ factor of human T-cell leukemia virus type I dimerizes with transcription factors JunB and c-Jun and modulates their transcriptional activity. J. Biol. Chem. 27843620-43627. - PubMed
-
- Boam, D. S., I. Davidson, and P. Chambon. 1995. A TATA-less promoter containing binding sites for ubiquitous transcription factors mediates cell type-specific regulation of the gene for transcription enhancer factor-1 (TEF-1). J. Biol. Chem. 27019487-19494. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Miscellaneous

