Use of shotgun proteomics for the identification, confirmation, and correction of C. elegans gene annotations
- PMID: 18653799
- PMCID: PMC2556273
- DOI: 10.1101/gr.077644.108
Use of shotgun proteomics for the identification, confirmation, and correction of C. elegans gene annotations
Abstract
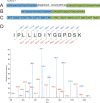
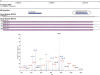
We describe a general mass spectrometry-based approach for gene annotation of any organism and demonstrate its effectiveness using the nematode Caenorhabditis elegans. We detected 6779 C. elegans proteins (67,047 peptides), including 384 that, although annotated in WormBase WS150, lacked cDNA or other prior experimental support. We also identified 429 new coding sequences that were unannotated in WS150. Nearly half (192/429) of the new coding sequences were confirmed with RT-PCR data. Thirty-three (approximately 8%) of the new coding sequences had been predicted to be pseudogenes, 151 (approximately 35%) reveal apparent errors in gene models, and 245 (57%) appear to be novel genes. In addition, we verified 6010 exon-exon splice junctions within existing WormBase gene models. Our work confirms that mass spectrometry is a powerful experimental tool for annotating sequenced genomes. In addition, the collection of identified peptides should facilitate future proteomics experiments targeted at specific proteins of interest.
Figures






References
-
- Anderson L., Hunter C.L., Hunter C.L. Quantitative mass spectrometric multiple reaction monitoring assays for major plasma proteins. Mol. Cell. Proteomics. 2006;5:573–588. - PubMed
-
- Basrai M.A., Hieter P., Boeke J.D., Hieter P., Boeke J.D., Boeke J.D. Small open reading frames: Beautiful needles in the haystack. Genome Res. 1997;7:768–771. - PubMed
-
- Brunner E., Ahrens C.H., Mohanty S., Baetschmann H., Loevenich S., Potthast F., Deutsch E.W., Panse C., de Lichtenberg U., Rinner O., Ahrens C.H., Mohanty S., Baetschmann H., Loevenich S., Potthast F., Deutsch E.W., Panse C., de Lichtenberg U., Rinner O., Mohanty S., Baetschmann H., Loevenich S., Potthast F., Deutsch E.W., Panse C., de Lichtenberg U., Rinner O., Baetschmann H., Loevenich S., Potthast F., Deutsch E.W., Panse C., de Lichtenberg U., Rinner O., Loevenich S., Potthast F., Deutsch E.W., Panse C., de Lichtenberg U., Rinner O., Potthast F., Deutsch E.W., Panse C., de Lichtenberg U., Rinner O., Deutsch E.W., Panse C., de Lichtenberg U., Rinner O., Panse C., de Lichtenberg U., Rinner O., de Lichtenberg U., Rinner O., Rinner O., et al. A high-quality catalog of the Drosophila melanogaster proteome. Nat. Biotechnol. 2007;25:576–583. - PubMed
-
- The C. elegans Sequencing Consortium Genome sequence of the nematode C. elegans: A platform for investigating biology. Science. 1998;282:2012–2018. - PubMed
-
- Desiere F., Deutsch E.W., King N.L., Nesvizhskii A.I., Mallick P., Eng J., Chen S., Eddes J., Loevenich S.N., Aebersold R., Deutsch E.W., King N.L., Nesvizhskii A.I., Mallick P., Eng J., Chen S., Eddes J., Loevenich S.N., Aebersold R., King N.L., Nesvizhskii A.I., Mallick P., Eng J., Chen S., Eddes J., Loevenich S.N., Aebersold R., Nesvizhskii A.I., Mallick P., Eng J., Chen S., Eddes J., Loevenich S.N., Aebersold R., Mallick P., Eng J., Chen S., Eddes J., Loevenich S.N., Aebersold R., Eng J., Chen S., Eddes J., Loevenich S.N., Aebersold R., Chen S., Eddes J., Loevenich S.N., Aebersold R., Eddes J., Loevenich S.N., Aebersold R., Loevenich S.N., Aebersold R., Aebersold R. The PeptideAtlas project. Nucleic Acids Res. 2006;34:D655–D658. - PMC - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
