An Argonaute transports siRNAs from the cytoplasm to the nucleus
- PMID: 18653886
- PMCID: PMC2771369
- DOI: 10.1126/science.1157647
An Argonaute transports siRNAs from the cytoplasm to the nucleus
Erratum in
- Science. 2009 Dec 4;326(5958):1346
Abstract
Ribonucleoprotein complexes consisting of Argonaute-like proteins and small regulatory RNAs function in a wide range of biological processes. Many of these small regulatory RNAs are predicted to act, at least in part, within the nucleus. We conducted a genetic screen to identify factors essential for RNA interference (RNAi) in nuclei of Caenorhabditis elegans and identified the Argonaute protein NRDE-3. In the absence of small interfering RNAs (siRNAs), NRDE-3 resides in the cytoplasm. NRDE-3 binds siRNAs generated by RNA-dependent RNA polymerases acting on messenger RNA templates in the cytoplasm and redistributes to the nucleus. Nuclear redistribution of NRDE-3 requires a functional nuclear localization signal, is required for nuclear RNAi, and results in NRDE-3 association with nuclear-localized nascent transcripts. Thus, specific Argonaute proteins can transport specific classes of small regulatory RNAs to distinct cellular compartments to regulate gene expression.
Figures
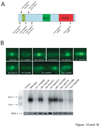

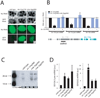
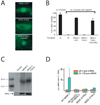
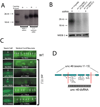
Comment in
-
Molecular biology. RNA interference in the nucleus.Science. 2008 Jul 25;321(5888):496-7. doi: 10.1126/science.1161854. Science. 2008. PMID: 18653868 No abstract available.
Similar articles
-
Small regulatory RNAs inhibit RNA polymerase II during the elongation phase of transcription.Nature. 2010 Jun 24;465(7301):1097-101. doi: 10.1038/nature09095. Epub 2010 Jun 13. Nature. 2010. PMID: 20543824 Free PMC article.
-
The Nrde Pathway Mediates Small-RNA-Directed Histone H3 Lysine 27 Trimethylation in Caenorhabditis elegans.Curr Biol. 2015 Sep 21;25(18):2398-403. doi: 10.1016/j.cub.2015.07.051. Epub 2015 Sep 10. Curr Biol. 2015. PMID: 26365259
-
Molecular biology. RNA interference in the nucleus.Science. 2008 Jul 25;321(5888):496-7. doi: 10.1126/science.1161854. Science. 2008. PMID: 18653868 No abstract available.
-
A new layer of rRNA regulation by small interference RNAs and the nuclear RNAi pathway.RNA Biol. 2017 Nov 2;14(11):1492-1498. doi: 10.1080/15476286.2017.1341034. Epub 2017 Jul 21. RNA Biol. 2017. PMID: 28640690 Free PMC article. Review.
-
Biology and Mechanisms of Short RNAs in Caenorhabditis elegans.Adv Genet. 2013;83:1-69. doi: 10.1016/B978-0-12-407675-4.00001-8. Adv Genet. 2013. PMID: 23890211 Review.
Cited by
-
RNAi pathways contribute to developmental history-dependent phenotypic plasticity in C. elegans.RNA. 2013 Mar;19(3):306-19. doi: 10.1261/rna.036418.112. Epub 2013 Jan 17. RNA. 2013. PMID: 23329696 Free PMC article.
-
Identification of small RNA pathway genes using patterns of phylogenetic conservation and divergence.Nature. 2013 Jan 31;493(7434):694-8. doi: 10.1038/nature11779. Epub 2012 Dec 23. Nature. 2013. PMID: 23364702 Free PMC article.
-
Gene silencing and Polycomb group proteins: an overview of their structure, mechanisms and phylogenetics.OMICS. 2013 Jun;17(6):283-96. doi: 10.1089/omi.2012.0105. OMICS. 2013. PMID: 23692361 Free PMC article. Review.
-
CDE-1 suppresses the production of risiRNA by coupling polyuridylation and degradation of rRNA.BMC Biol. 2020 Sep 4;18(1):115. doi: 10.1186/s12915-020-00850-z. BMC Biol. 2020. PMID: 32887607 Free PMC article.
-
Human tRNA-derived small RNAs in the global regulation of RNA silencing.RNA. 2010 Apr;16(4):673-95. doi: 10.1261/rna.2000810. Epub 2010 Feb 24. RNA. 2010. PMID: 20181738 Free PMC article.
References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

