A patterned anisotropic nanofluidic sieving structure for continuous-flow separation of DNA and proteins
- PMID: 18654231
- PMCID: PMC2621439
- DOI: 10.1038/nnano.2006.206
A patterned anisotropic nanofluidic sieving structure for continuous-flow separation of DNA and proteins
Abstract
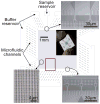
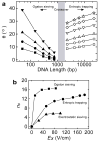
Microfabricated regular sieving structures hold great promise as an alternative to gels to improve the speed and resolution of biomolecule separation. In contrast to disordered porous gel networks, these regular structures also provide well defined environments ideal for the study of molecular dynamics in confining spaces. However, the use of regular sieving structures has, to date, been limited to the separation of long DNA molecules, however separation of smaller, physiologically relevant macromolecules, such as proteins, still remains a challenge. Here we report a microfabricated anisotropic sieving structure consisting of a two-dimensional periodic nanofluidic filter array. The designed structural anisotropy causes different-sized or -charged biomolecules to follow distinct trajectories, leading to efficient separation. Continuous-flow size-based separation of DNA and proteins, as well as electrostatic separation of proteins, was achieved, demonstrating the potential use of this device as a generic molecular sieving structure for an integrated biomolecule sample preparation and analysis system.
Figures




Comment in
-
Nanofluidics: a fork in the nano-road.Nat Nanotechnol. 2007 Feb;2(2):79-80. doi: 10.1038/nnano.2007.18. Nat Nanotechnol. 2007. PMID: 18654220 No abstract available.
References
-
- Scopes RK. Protein Purification, Principles and Practice. 3. Springer-Verlag; New York: 1993.
-
- Giddings JC. Principles and Theory. Marcel Dekker; New York: 1965. Dynamics of Chromatography. Part 1.
-
- Slater GW, Mayer P, Drouin G. Migration of DNA through gels. Methods Enzymol. 1996;270:272–295. - PubMed
-
- Viovy JL. Electrophoresis of DNA and other polyelectrolytes: physical mechanisms. Rev Mod Phys. 2000;72:813–872.
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

