Sequence-specific detection of individual DNA polymerase complexes in real time using a nanopore
- PMID: 18654412
- PMCID: PMC2507869
- DOI: 10.1038/nnano.2007.344
Sequence-specific detection of individual DNA polymerase complexes in real time using a nanopore
Abstract
Nanoscale pores have potential to be used as biosensors and are an established tool for analysing the structure and composition of single DNA or RNA molecules. Recently, nanopores have been used to measure the binding of enzymes to their DNA substrates. In this technique, a polynucleotide bound to an enzyme is drawn into the nanopore by an applied voltage. The force exerted on the charged backbone of the polynucleotide by the electric field is used to examine the enzyme-polynucleotide interactions. Here we show that a nanopore sensor can accurately identify DNA templates bound in the catalytic site of individual DNA polymerase molecules. Discrimination among unbound DNA, binary DNA/polymerase complexes, and ternary DNA/polymerase/deoxynucleotide triphosphate complexes was achieved in real time using finite state machine logic. This technique is applicable to numerous enzymes that bind or modify DNA or RNA including exonucleases, kinases and other polymerases.
Figures
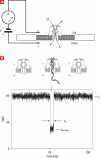

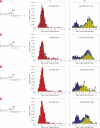
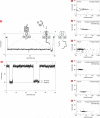

References
-
- Deamer DW, Branton D. Characterization of nucleic acids by nanopore analysis. Acc. Chem. Res. 2002;35:817–825. - PubMed
-
- Dekker C. Solid-state nanopores. Nature Nanotechnol. 2007;2:209–215. - PubMed
-
- Siwy Z, et al. Protein biosensors based on biofunctionalized conical gold nanotubes. J. Am. Chem. Soc. 2005;127:5000–5001. - PubMed
-
- Hornblower B, et al. Single-molecule analysis of DNA–protein complexes using nanopores. Nature Methods. 2007;4:315–317. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

