Recapturing and trapping single molecules with a solid-state nanopore
- PMID: 18654430
- PMCID: PMC3174059
- DOI: 10.1038/nnano.2007.381
Recapturing and trapping single molecules with a solid-state nanopore
Abstract
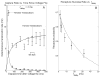
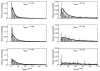
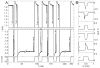
The development of solid-state nanopores, inspired by their biological counterparts, shows great potential for the study of single macromolecules. Applications such as DNA sequencing and the exploration of protein folding require control of the dynamics of the molecule's interaction with the pore, but DNA capture by a solid-state nanopore is not well understood. By recapturing individual molecules soon after they pass through a nanopore, we reveal the mechanism by which double-stranded DNA enters the pore. The observed recapture rates and times agree with solutions of a drift-diffusion model. Electric forces draw DNA to the pore over micrometer-scale distances, and upon arrival at the pore, molecules begin translocation almost immediately. Repeated translocation of the same molecule improves measurement accuracy, offers a way to probe the chemical transformations and internal dynamics of macromolecules on sub-millisecond time and sub-micrometre length scales, and demonstrates the ability to trap, study and manipulate individual macromolecules in solution.
Figures

References
-
- Li J, et al. Ion-beam sculpting at nanometre length scales. Nature. 2001;412:166–169. - PubMed
-
- Storm AJ, Chen JH, Ling XS, Zandbergen HW, Dekker C. Electron-beam-induced deformations of SiO2 nanostructures. Journal of Applied Physics. 2005;98
-
- Wu MY, Krapf D, Zandbergen M, Zandbergen H, Batson PE. Formation of nanopores in a SiN/SiO2 membrane with an electron beam. Applied Physics Letters. 2005;87
-
- Lo CJ, Aref T, Bezryadin A. Fabrication of symmetric sub-5 nm nanopores using focused ion and electron beams. Nanotechnology. 2006;17:3264–3267.
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

