The linker domain of poly(rC) binding protein 2 is a major determinant in poliovirus cap-independent translation
- PMID: 18656221
- PMCID: PMC2595138
- DOI: 10.1016/j.virol.2008.05.007
The linker domain of poly(rC) binding protein 2 is a major determinant in poliovirus cap-independent translation
Abstract
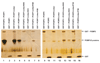
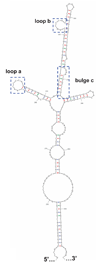
Poliovirus, a member of the enterovirus genus in the family Picornaviridae, is the causative agent of poliomyelitis. Translation of the viral genome is mediated through an internal ribosomal entry site (IRES) encoded within the 5' noncoding region (5' NCR). IRES elements are highly structured RNA sequences that facilitate the recruitment of ribosomes for translation. Previous studies have shown that binding of a cellular protein, poly(rC) binding protein 2 (PCBP2), to a major stem-loop structure in the genomic 5' NCR is necessary for the translation of picornaviruses containing type I IRES elements, including poliovirus, coxsackievirus, and human rhinovirus. PCBP1, an isoform that shares approximately 90% amino acid identity to PCBP2, cannot efficiently stimulate poliovirus IRES-mediated translation, most likely due to its reduced binding affinity to stem-loop IV within the poliovirus IRES. The primary differences between PCBP1 and PCBP2 are found in the so-called linker domain between the second and third K-homology (KH) domains of these proteins. We hypothesize that the linker region of PCBP2 augments binding to poliovirus stem-loop IV RNA. To test this hypothesis, we generated six PCBP1/PCBP2 chimeric proteins. The recombinant PCBP1/PCBP2 chimeric proteins were able to interact with poliovirus stem-loop I RNA and participate in protein-protein interactions. We demonstrated that the PCBP1/PCBP2 chimeric proteins with the PCBP2 linker, but not with the PCBP1 linker, were able to interact with poliovirus stem-loop IV RNA, and could subsequently stimulate poliovirus IRES-mediated translation. In addition, using a monoclonal anti-PCBP2 antibody (directed against the PCBP2 linker domain) in mobility shift assays, we showed that the PCBP2 linker domain modulates binding to poliovirus stem-loop IV RNA via a mechanism that is not inhibited by the antibody.
Figures









References
-
- Andino R, Rieckhof GE, Baltimore D. A functional ribonucleoprotein complex forms around the 5' end of poliovirus RNA. Cell. 1990;63:369–380. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous

