Rapid "open-source" engineering of customized zinc-finger nucleases for highly efficient gene modification
- PMID: 18657511
- PMCID: PMC2535758
- DOI: 10.1016/j.molcel.2008.06.016
Rapid "open-source" engineering of customized zinc-finger nucleases for highly efficient gene modification
Abstract
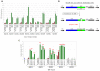
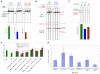
Custom-made zinc-finger nucleases (ZFNs) can induce targeted genome modifications with high efficiency in cell types including Drosophila, C. elegans, plants, and humans. A bottleneck in the application of ZFN technology has been the generation of highly specific engineered zinc-finger arrays. Here we describe OPEN (Oligomerized Pool ENgineering), a rapid, publicly available strategy for constructing multifinger arrays, which we show is more effective than the previously published modular assembly method. We used OPEN to construct 37 highly active ZFN pairs which induced targeted alterations with high efficiencies (1%-50%) at 11 different target sites located within three endogenous human genes (VEGF-A, HoxB13, and CFTR), an endogenous plant gene (tobacco SuRA), and a chromosomally integrated EGFP reporter gene. In summary, OPEN provides an "open-source" method for rapidly engineering highly active zinc-finger arrays, thereby enabling broader practice, development, and application of ZFN technology for biological research and gene therapy.
Figures


References
-
- Alwin S, Gere MB, Guhl E, Effertz K, Barbas CF, 3rd, Segal DJ, Weitzman MD, Cathomen T. Custom zinc-finger nucleases for use in human cells. Mol Ther. 2005;12:610–617. - PubMed
-
- Bae KH, Do Kwon Y, Shin HC, Hwang MS, Ryu EH, Park KS, Yang HY, Lee DK, Lee Y, Park J, et al. Human zinc fingers as building blocks in the construction of artificial transcription factors. Nat Biotechnol. 2003;21:275–280. - PubMed
-
- Beerli RR, Barbas CF., 3rd Engineering polydactyl zinc-finger transcription factors. Nat Biotechnol. 2002;20:135–141. - PubMed
-
- Bibikova M, Beumer K, Trautman JK, Carroll D. Enhancing gene targeting with designed zinc finger nucleases. Science. 2003;300:764. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials

