Identification and characterization of bacterial cutinase
- PMID: 18658138
- PMCID: PMC3258855
- DOI: 10.1074/jbc.M800848200
Identification and characterization of bacterial cutinase
Abstract
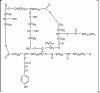
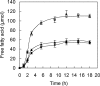
Cutinase, which exists in both fungi and bacteria, catalyzes the cleavage of the ester bonds of cutin. Fungal cutinases have been extensively studied, however, reports on bacterial cutinases have been limited due to the lack of knowledge concerning the identity of their open reading frames. In the present study, the cutinase from Thermobifida fusca was induced by cutin and purified to homogeneity by following p-nitrophenyl butyrate hydrolyzing activity. Peptide mass fingerprinting analysis of the wild-type enzyme matched two proteins, Tfu_0883 and Tfu_0882, which are 93% identical in sequence. Both proteins were cloned and overexpressed in their mature form. Recombinant Tfu_0883 and Tfu_0882 display very similar enzymatic properties and were confirmed to be cutinases by their capability to hydrolyze the ester bonds of cutin. Comparative characterization of Fusarium solani pisi and T. fusca cutinases indicated that they have similar substrate specificity and catalytic properties except that the T. fusca enzymes are thermally more stable. Homology modeling revealed that T. fusca cutinases adopt an alpha/beta-hydrolase fold that exhibits both similarities and variations from the fungal cutinase structure. A serine hydrolase catalytic mechanism involving a Ser(170)-His(248)-Asp(216) (Tfu_0883 numbering) catalytic triad was supported by active site-directed inhibition studies and mutational analyses. This is the first report of cutinase encoding genes from bacterial sources.
Figures





References
-
- Walton, T. J., and Kolattukudy, P. E. (1972) Biochemistry 11 1885–1896 - PubMed
-
- Purdy, R. E., and Kolattukudy, P. E. (1973) Arch. Biochem. Biophys. 159 61–69 - PubMed
-
- Gerard, H. C., Osman, S. F., Fett, W. F., and Moreau, R. A. (1992) Phytochem. Analysis 3 139–144
-
- Ferreira, B. S., Calado, C. R., van Keulen, F., Fonseca, L. P., Cabral, J. M., and da Fonseca, M. M. (2003) Appl. Microbiol. Biotechnol. 61 69–76 - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources

