SNAP-25 substrate peptide (residues 180-183) binds to but bypasses cleavage by catalytically active Clostridium botulinum neurotoxin E
- PMID: 18658150
- PMCID: PMC2533769
- DOI: 10.1074/jbc.M803756200
SNAP-25 substrate peptide (residues 180-183) binds to but bypasses cleavage by catalytically active Clostridium botulinum neurotoxin E
Abstract
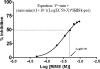
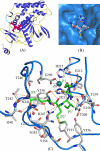
Clostridium botulinum neurotoxins are the most potent toxins to humans. The recognition and cleavage of SNAREs are prime evente in exhibiting their toxicity. We report here the crystal structure of the catalytically active full-length botulinum serotype E catalytic domain (BoNT E) in complex with SNAP-25 (a SNARE protein) substrate peptide Arg(180)-Ile(181)-Met(182)-Glu(183) (P1-P3'). It is remarkable that the peptide spanning the scissile bond binds to but bypasses cleavage by the enzyme and inhibits the catalysis fairly with K(i) approximately 69 microm. The inhibitory peptide occupies the active site of BoNT E and shows well defined electron density. The catalytic zinc and the conserved key residue Tyr(350) of the enzyme facilitate the docking of Arg(180) (P1) by interacting with its carbonyl oxygen that displaces the nucleophilic water. The general base Glu(212) side chain interacts with the main chain amino group of P1 and P1'. Conserved Arg(347) of BoNT E stabilizes the proper docking of the Ile(181) (P1') main chain, whereas the hydrophobic pockets stabilize the side chains of Ile(181) (P1') and Met(182) (P2'), and the 250 loop stabilizes Glu(183) (P3'). Structural and functional analysis revealed an important role for the P1' residue and S1' pocket in driving substrate recognition and docking at the active site. This study is the first of its kind and rationalizes the substrate cleavage strategy of BoNT E. Also, our complex structure opens up an excellent opportunity of structure-based drug design for this fast acting and extremely toxic high priority BoNT E.
Figures



References
-
- DasGupta, B. R., and Rasmussen, S. (1983) Toxicon 21 566–569 - PubMed
-
- Dasgupta, B. R., and Rasmussen, S. (1983) Toxicon 21 535–545 - PubMed
-
- Sathyamoorthy, V., and DasGupta, B. R. (1985) Biochem. Biophys. Res. Commun. 127 768–772 - PubMed
-
- Sathyamurthy, V., and Dasgupta, B. R. (1985) J. Biol. Chem. 260 10461–10466 - PubMed
-
- Montecucco, C., and Schiavo, G. (1994) Mol. Microbiol. 13 1–9 - PubMed
Publication types
MeSH terms
Substances
Associated data
- Actions
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous

