Characterization of the control catabolite protein of gluconeogenic genes repressor by fluorescence cross-correlation spectroscopy and other biophysical approaches
- PMID: 18658229
- PMCID: PMC2567959
- DOI: 10.1529/biophysj.108.135863
Characterization of the control catabolite protein of gluconeogenic genes repressor by fluorescence cross-correlation spectroscopy and other biophysical approaches
Erratum in
- Biophys J. 2009 Feb;96(3):1232
Abstract
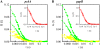
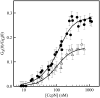
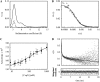
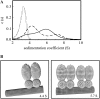
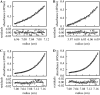
Determination of the physical parameters underlying protein-DNA interactions is crucial for understanding the regulation of gene expression. In particular, knowledge of the stoichiometry of the complexes is a prerequisite to determining their energetics and functional molecular mechanisms. However, the experimental determination of protein-DNA complex stoichiometries remains challenging. We used fluorescence cross-correlation spectroscopy (FCCS) to investigate the interactions of the control catabolite protein of gluconeogenic genes, a key metabolic regulator in Gram-positive bacteria, with two oligonucleotides derived from its target operator sequences, gapB and pckA. According to our FCCS experiments, the stoichiometry of binding is twofold larger for the pckA target than for gapB. Correcting the FCCS data for protein self-association indicated that control catabolite protein of gluconeogenic genes forms dimeric complexes on the gapB target and tetrameric complexes on the pckA target. Analytical ultracentrifugation coupled with fluorescence anisotropy and hydrodynamic modeling allowed unambiguous confirmation of this result. The use of multiple complementary techniques to characterize these complexes should be employed wherever possible. However, there are cases in which analytical ultracentrifugation is precluded, due to protein stability, solubility, or availability, or, more obviously, when the studies are carried out in live cells. If information concerning the self-association of the protein is available, FCCS can be used for the direct and simultaneous determination of the affinity, cooperativity, and stoichiometry of protein-DNA complexes in a concentration range and conditions relevant to the regulation of these interactions.
Figures
References
-
- Hwang, L., and T. Wohland. 2004. Dual-color fluorescence cross-correlation spectroscopy using single laser wavelength excitation. ChemPhysChem. 5:549–551. - PubMed
-
- Rippe, K. 2000. Simultaneous binding of two DNA duplexes to the NtrC-enhancer complex studied by two-color fluorescence cross-correlation spectroscopy. Biochemistry. 39:2131–2139. - PubMed
-
- Strohner, R., M. Wachsmuth, K. Dachauer, J. Mazurkiewicz, J. Hochstatter, K. Rippe, and G. Langst. 2005. A ‘loop recapture’ mechanism for ACF-dependent nucleosome remodeling. Nat. Struct. Mol. Biol. 12:683–690. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources

