60S ribosomal subunit assembly dynamics defined by semi-quantitative mass spectrometry of purified complexes
- PMID: 18658244
- PMCID: PMC2528192
- DOI: 10.1093/nar/gkn469
60S ribosomal subunit assembly dynamics defined by semi-quantitative mass spectrometry of purified complexes
Abstract
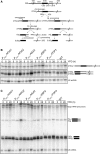
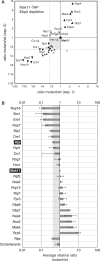
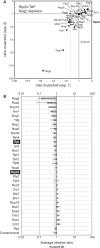
During the highly conserved process of eukaryotic ribosome formation, RNA follows a maturation path with well-defined, successive intermediates that dynamically associate with many pre-ribosomal proteins. A comprehensive description of the assembly process is still lacking. To obtain data on the timing and order of association of the different pre-ribosomal factors, a strategy consists in the use of pre-ribsomal particles isolated from mutants that block ribosome formation at different steps. Immunoblots, inherently limited to only a few factors, have been applied to evaluate the accumulation or decrease of pre-ribosomal intermediates under mutant conditions. For a global protein-level description of different 60S ribosomal subunit maturation intermediates in yeast, we have adapted a method of in vivo isotopic labelling and mass spectrometry to study pre-60S complexes isolated from strains in which rRNA processing was affected by individual depletion of five factors: Ebp2, Nog1, Nsa2, Nog2 or Pop3. We obtained quantitative data for 45 distinct pre-60S proteins and detected coordinated changes for over 30 pre-60S factors in the analysed mutants. These results led to the characterisation of the composition of early, intermediate and late pre-ribosomal complexes, specific for crucial maturation steps during 60S assembly in eukaryotes.
Figures




References
-
- Ferreira-Cerca S, Poll G, Kuhn H, Neueder A, Jakob S, Tschochner H, Milkereit P. Analysis of the in vivo assembly pathway of eukaryotic 40S ribosomal proteins. Mol. Cell. 2007;28:446–457. - PubMed
-
- Fromont-Racine M, Senger B, Saveanu C, Fasiolo F. Ribosome assembly in eukaryotes. Gene. 2003;313:17–42. - PubMed
-
- Tschochner H, Hurt E. Pre-ribosomes on the road from the nucleolus to the cytoplasm. Trends Cell Biol. 2003;13:255–263. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

