Selective recognition of pyrimidine-pyrimidine DNA mismatches by distance-constrained macrocyclic bis-intercalators
- PMID: 18658249
- PMCID: PMC2528167
- DOI: 10.1093/nar/gkn392
Selective recognition of pyrimidine-pyrimidine DNA mismatches by distance-constrained macrocyclic bis-intercalators
Abstract
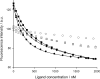
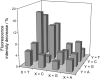
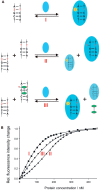
Binding of three macrocyclic bis-intercalators, derivatives of acridine and naphthalene, and two acyclic model compounds to mismatch-containing and matched duplex oligodeoxynucleotides was analyzed by thermal denaturation experiments, electrospray ionization mass spectrometry studies (ESI-MS) and fluorescent intercalator displacement (FID) titrations. The macrocyclic bis-intercalators bind to duplexes containing mismatched thymine bases with high selectivity over the fully matched ones, whereas the acyclic model compounds are much less selective and strongly bind to the matched DNA. Moreover, the results from thermal denaturation experiments are in very good agreement with the binding affinities obtained by ESI-MS and FID measurements. The FID results also demonstrate that the macrocyclic naphthalene derivative BisNP preferentially binds to pyrimidine-pyrimidine mismatches compared to all other possible base mismatches. This ligand also efficiently competes with a DNA enzyme (M.TaqI) for binding to a duplex with a TT-mismatch, as shown by competitive fluorescence titrations. Altogether, our results demonstrate that macrocyclic distance-constrained bis-intercalators are efficient and selective mismatch-binding ligands that can interfere with mismatch-binding enzymes.
Figures
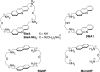






Similar articles
-
Recognition of homopyrimidine mismatches by distance-constrained macrocyclic bisintercalators.Nucleic Acids Symp Ser (Oxf). 2008;(52):109-10. doi: 10.1093/nass/nrn056. Nucleic Acids Symp Ser (Oxf). 2008. PMID: 18776277
-
Copper(II)-Controlled Molecular Glue for Mismatched DNA.Chembiochem. 2017 Apr 4;18(7):618-622. doi: 10.1002/cbic.201600675. Epub 2017 Feb 20. Chembiochem. 2017. PMID: 28106332
-
Macrocyclic DNA-mismatch-binding ligands: structural determinants of selectivity.Chemistry. 2010 Jan 18;16(3):878-89. doi: 10.1002/chem.200901989. Chemistry. 2010. PMID: 19938008
-
The search of DNA-intercalators as antitumoral drugs: what it worked and what did not work.Curr Med Chem. 2005;12(2):127-51. doi: 10.2174/0929867053363414. Curr Med Chem. 2005. PMID: 15638732 Review.
-
The interaction of intercalators and groove-binding agents with DNA triple-helical structures: the influence of ligand structure, DNA backbone modifications and sequence.J Mol Recognit. 1994 Jun;7(2):89-98. doi: 10.1002/jmr.300070206. J Mol Recognit. 1994. PMID: 7826678 Review.
Cited by
-
Investigation on the Interactions of NiCR and NiCR-2H with DNA.Bioinorg Chem Appl. 2010;2010:619436. doi: 10.1155/2010/619436. Epub 2010 Jun 30. Bioinorg Chem Appl. 2010. PMID: 20671951 Free PMC article.
-
Differential targeting of unpaired bases within duplex DNA by the natural compound clerocidin: a valuable tool to dissect DNA secondary structure.PLoS One. 2012;7(12):e52994. doi: 10.1371/journal.pone.0052994. Epub 2012 Dec 28. PLoS One. 2012. PMID: 23285245 Free PMC article.
-
NMR determination of the 2:1 binding complex of naphthyridine carbamate dimer (NCD) and CGG/CGG triad in double-stranded DNA.Nucleic Acids Res. 2022 Sep 23;50(17):9621-9631. doi: 10.1093/nar/gkac740. Nucleic Acids Res. 2022. PMID: 36095126 Free PMC article.
-
Ratiometric Detection of ATP by Fluorescent Cyclophanes with Bellows-Type Sensing Mechanism.Chemistry. 2020 Aug 6;26(44):9991-9997. doi: 10.1002/chem.202001523. Epub 2020 Jul 16. Chemistry. 2020. PMID: 32497327 Free PMC article.
-
May the Best Molecule Win: Competition ESI Mass Spectrometry.Int J Mol Sci. 2015 Oct 15;16(10):24506-31. doi: 10.3390/ijms161024506. Int J Mol Sci. 2015. PMID: 26501262 Free PMC article. Review.
References
-
- Echols H, Goodman MF. Fidelity mechanisms in DNA replication. Annu. Rev. Biochem. 1991;60:477–511. - PubMed
-
- Mendelman LV, Boosalis MS, Petruska J, Goodman MF. Nearest neighbor influences on DNA polymerase insertion fidelity. J. Biol. Chem. 1989;264:14415–14423. - PubMed
-
- Lee AM, Xiao J, Singleton SF. Origins of sequence selectivity in homologous genetic recombination: insights from rapid kinetic probing of RecA-mediated DNA strand exchange. J. Mol. Biol. 2006;360:343–359. - PubMed
-
- Gacy AM, Goellner G, Juranic N, Macura S, McMurray CT. Trinucleotide repeats that expand in human disease form hairpin structures in vitro. Cell. 1995;81:533–540. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Miscellaneous

