Dispersed mutations in histone H3 that affect transcriptional repression and chromatin structure of the CHA1 promoter in Saccharomyces cerevisiae
- PMID: 18658255
- PMCID: PMC2568054
- DOI: 10.1128/EC.00233-08
Dispersed mutations in histone H3 that affect transcriptional repression and chromatin structure of the CHA1 promoter in Saccharomyces cerevisiae
Abstract
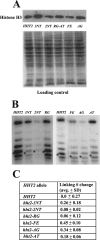
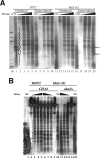
The histone H3 amino terminus, but not that of H4, is required to prevent the constitutively bound activator Cha4 from remodeling chromatin and activating transcription at the CHA1 gene in Saccharomyces cerevisiae. Here we show that neither the modifiable lysine residues nor any specific region of the H3 tail is required for repression of CHA1. We then screened for histone H3 mutations that cause derepression of the uninduced CHA1 promoter and identified six mutants, three of which are also temperature-sensitive mutants and four of which exhibit a sin(-) phenotype. Histone mutant levels were similar to that of wild-type H3, and the mutations did not cause gross alterations in nucleosome structure. One specific and strongly derepressing mutation, H3 A111G, was examined in depth and found to cause a constitutively active chromatin configuration at the uninduced CHA1 promoter as well as at the ADH2 promoter. Transcriptional derepression and altered chromatin structure of the CHA1 promoter depend on the activator Cha4. These results indicate that modest perturbations in distinct regions of the nucleosome can substantially affect the repressive function of chromatin, allowing activation in the absence of a normal inducing signal (at CHA1) or of Swi/Snf (resulting in a sin(-) phenotype).
Figures
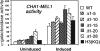






References
-
- Berger, S. L. 2007. The complex language of chromatin regulation during transcription. Nature 447407-412. - PubMed
-
- Christianson, T. W., R. S. Sikorski, M. Dante, J. H. Shero, and P. Hieter. 1992. Multifunctional yeast high-copy-number shuttle vectors. Gene 110119-122. - PubMed
-
- Cosgrove, M. S., J. D. Boeke, and C. Wolberger. 2004. Regulated nucleosome mobility and the histone code. Nat. Struct. Mol. Biol. 111037-1043. - PubMed
Publication types
MeSH terms
Substances
Associated data
- Actions
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

