The EPIP peptide of INFLORESCENCE DEFICIENT IN ABSCISSION is sufficient to induce abscission in arabidopsis through the receptor-like kinases HAESA and HAESA-LIKE2
- PMID: 18660431
- PMCID: PMC2518227
- DOI: 10.1105/tpc.108.059139
The EPIP peptide of INFLORESCENCE DEFICIENT IN ABSCISSION is sufficient to induce abscission in arabidopsis through the receptor-like kinases HAESA and HAESA-LIKE2
Abstract
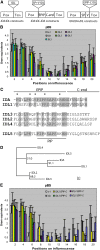
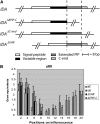
In Arabidopsis thaliana, the final step of floral organ abscission is regulated by INFLORESCENCE DEFICIENT IN ABSCISSION (IDA): ida mutants fail to abscise floral organs, and plants overexpressing IDA display earlier abscission. We show that five IDA-LIKE (IDL) genes are expressed in different tissues, but plants overexpressing these genes have phenotypes similar to IDA-overexpressing plants, suggesting functional redundancy. IDA/IDL proteins have N-terminal signal peptides and a C-terminal conserved motif (extended PIP [EPIP]) at the C terminus (EPIP-C). IDA can, similar to CLAVATA3, be processed by an activity from cauliflower meristems. The EPIP-C of IDA and IDL1 replaced IDA function in vivo, when the signal peptide was present. In addition, synthetic IDA and IDL1 EPIP peptides rescued ida and induced early floral abscission in wild-type flowers. The EPIP-C of the other IDL proteins could partially substitute for IDA function. Similarly to ida, a double mutant between the receptor-like kinases (RLKs) HAESA (HAE) and HAESA-LIKE2 (HSL2) displays nonabscising flowers. Neither overexpression of IDA nor synthetic EPIP or EPIP-C peptides could rescue the hae hsl2 abscission deficiency. We propose that IDA and the IDL proteins constitute a family of putative ligands that act through RLKs to regulate different events during plant development.
Figures




References
-
- Aalen, R.B., Butenko, M.A., Stenvik, G.E., Tandstad, N.M., and Patterson, S.E. (2006). Genetic control of floral abscission. In Floriculture, Ornamental and Plant Biotechnology: Advances and Topical Issues J.T. de Silva, ed (London: Global Science Books), pp. 101–108.
-
- Alonso, J.M., et al. (2003). Genome-wide insertional mutagenesis of Arabidopsis thaliana. Science 301 653–657. - PubMed
-
- Butenko, M.A., Stenvik, G.E., Alm, V., Sæther, B., Patterson, S.E., and Aalen, R.B. (2006). Ethylene-dependent and -independent pathways controlling floral abscission are revealed to converge using promoter:reporter gene constructs in the ida abscission mutant. J. Exp. Bot. 57 3627–3637. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials

