Abnormal liver development and resistance to 2,3,7,8-tetrachlorodibenzo-p-dioxin toxicity in mice carrying a mutation in the DNA-binding domain of the aryl hydrocarbon receptor
- PMID: 18660548
- PMCID: PMC2563146
- DOI: 10.1093/toxsci/kfn149
Abnormal liver development and resistance to 2,3,7,8-tetrachlorodibenzo-p-dioxin toxicity in mice carrying a mutation in the DNA-binding domain of the aryl hydrocarbon receptor
Abstract
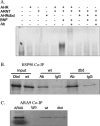
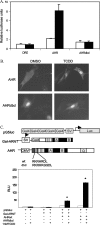
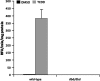
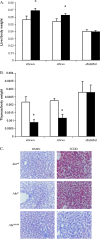
The aryl hydrocarbon receptor (AHR) is known for its role in the adaptive and toxic responses to a large number of environmental contaminants, as well as its role in hepatovascular development. The classical AHR pathway involves ligand binding, nuclear translocation, heterodimerization with the AHR nuclear translocator (ARNT), and binding of the heterodimer to dioxin response elements (DREs), thereby modulating the transcription of an array of genes. The AHR has also been implicated in signaling events independent of nuclear localization and DNA binding, and it has been suggested that such pathways may play important roles in the toxicity of 2,3,7,8-tetrachlorodibenzo-p-dioxin (TCDD). Here, we report the generation of a mouse model that expresses an AHR protein capable of ligand binding, interactions with chaperone proteins, functional heterodimerization with ARNT, and nuclear translocation, but is unable to bind DREs. Using this model, we provide evidence that DNA binding is required AHR-mediated liver development, as Ahr(dbd/dbd) mice exhibit a patent ductus venosus, similar to what is seen in Ahr(-/-) mice. Furthermore, Ahr(dbd/dbd) mice are resistant to TCDD-induced toxicity for all endpoints tested. These data suggest that DNA binding is necessary for AHR-mediated developmental and toxic signaling.
Figures



References
-
- Bacsi SG, Hankinson O. Functional characterization of DNA-binding domains of the subunits of the heterodimeric aryl hydrocarbon receptor complex imputing novel and canonical basic helix-loop-helix protein-DNA interactions. J. Biol. Chem. 1996;271:8843–8850. - PubMed
-
- Blankenship A, Matsumura F. 2,3,7,8-Tetrachlorodibenzo-p-dioxin-induced activation of a protein tyrosine kinase, pp60src, in murine hepatic cytosol using a cell-free system. Mol. Pharmacol. 1997;52:667–675. - PubMed
-
- Bunger MK, Moran SM, Glover E, Thomae TL, Lahvis GP, Lin BC, Bradfield CA. Resistance to 2,3,7,8-tetrachlorodibenzo-p-dioxin toxicity and abnormal liver development in mice carrying a mutation in the nuclear localization sequence of the aryl hydrocarbon receptor. J. Biol. Chem. 2003;278:17767–17774. - PubMed
-
- Buters JT, Doehmer J, Gonzalez FJ. Cytochrome P450-null mice. Drug Metab. Rev. 1999;31:437–447. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

