Novel splicing factor RBM25 modulates Bcl-x pre-mRNA 5' splice site selection
- PMID: 18663000
- PMCID: PMC2546994
- DOI: 10.1128/MCB.00560-08
Novel splicing factor RBM25 modulates Bcl-x pre-mRNA 5' splice site selection
Abstract
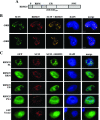
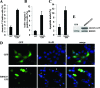
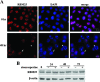
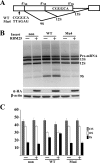
RBM25 has been shown to associate with splicing cofactors SRm160/300 and assembled splicing complexes, but little is known about its splicing regulation. Here, we characterize the functional role of RBM25 in alternative pre-mRNA splicing. Increased RBM25 expression correlated with increased apoptosis and specifically affected the expression of Bcl-x isoforms. RBM25 stimulated proapoptotic Bcl-x(S) 5' splice site (5' ss) selection in a dose-dependent manner, whereas its depletion caused the accumulation of antiapoptotic Bcl-x(L). Furthermore, RBM25 specifically bound to Bcl-x RNA through a CGGGCA sequence located within exon 2. Mutation in this element abolished the ability of RBM25 to enhance Bcl-x(S) 5' ss selection, leading to decreased Bcl-x(S) isoform expression. Binding of RBM25 was shown to promote the recruitment of the U1 small nuclear ribonucleoprotein particle (snRNP) to the weak 5' ss; however, it was not required when a strong consensus 5' ss was present. In support of a role for RBM25 in modulating the selection of a 5' ss, we demonstrated that RBM25 associated selectively with the human homolog of yeast U1 snRNP-associated factor hLuc7A. These data suggest a novel mode for Bcl-x(S) 5' ss activation in which binding of RBM25 with exonic element CGGGCA may stabilize the pre-mRNA-U1 snRNP through interactions with hLuc7A.
Figures






References
-
- Bingle, C. D., R. W. Craig, B. M. Swales, V. Singleton, P. Zhou, and M. K. Whyte. 2000. Exon skipping in Mcl-1 results in a bcl-2 homology domain 3 only gene product that promotes cell death. J. Biol. Chem. 27522136-22146. - PubMed
-
- Black, D. L. 2003. Mechanisms of alternative pre-messenger RNA splicing. Annu. Rev. Biochem. 72291-336. - PubMed
-
- Blencowe, B. J. 2000. Exonic splicing enhancers: mechanism of action, diversity and role in human genetic diseases. Trends Biochem. Sci. 25106-110. - PubMed
-
- Boise, L. H., M. Gonzalez-Garcia, C. E. Postema, L. Ding, T. Lindsten, L. A. Turka, X. Mao, G. Nunez, and C. B. Thompson. 1993. bcl-x, a bcl-2-related gene that functions as a dominant regulator of apoptotic cell death. Cell 74597-608. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials
