Extensive meiotic asynapsis in mice antagonises meiotic silencing of unsynapsed chromatin and consequently disrupts meiotic sex chromosome inactivation
- PMID: 18663141
- PMCID: PMC2483523
- DOI: 10.1083/jcb.200710195
Extensive meiotic asynapsis in mice antagonises meiotic silencing of unsynapsed chromatin and consequently disrupts meiotic sex chromosome inactivation
Abstract
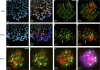
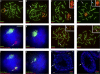
Chromosome synapsis during zygotene is a prerequisite for the timely homologous recombinational repair of meiotic DNA double-strand breaks (DSBs). Unrepaired DSBs are thought to trigger apoptosis during midpachytene of male meiosis if synapsis fails. An early pachytene response to asynapsis is meiotic silencing of unsynapsed chromatin (MSUC), which, in normal males, silences the X and Y chromosomes (meiotic sex chromosome inactivation [MSCI]). In this study, we show that MSUC occurs in Spo11-null mouse spermatocytes with extensive asynapsis but lacking meiotic DSBs. In contrast, three mutants (Dnmt3l, Msh5, and Dmc1) with high levels of asynapsis and numerous persistent unrepaired DSBs have a severely impaired MSUC response. We suggest that MSUC-related proteins, including the MSUC initiator BRCA1, are sequestered at unrepaired DSBs. All four mutants fail to silence the X and Y chromosomes (MSCI failure), which is sufficient to explain the midpachytene apoptosis. Apoptosis does not occur in mice with a single additional asynapsed chromosome with unrepaired meiotic DSBs and no disturbance of MSCI.
Figures






References
-
- Ahmed, E.A., A. van der Vaart, A. Barten, H.B. Kal, J. Chen, Z. Lou, K. Minter-Dykhouse, J. Bartkova, J. Bartek, P. de Boer, and D.G. de Rooij. 2007. Differences in DNA double strand breaks repair in male germ cell types: lessons learned from a differential expression of Mdc1 and 53BP1. DNA Repair (Amst.). 6:1243–1254. - PubMed
-
- Ashley, T., A.P. Gaeth, L.B. Creemers, A.M. Hack, and D.G. de Rooij. 2004. a. Correlation of meiotic events in testis sections and microspreads of mouse spermatocytes relative to the mid-pachytene checkpoint. Chromosoma. 113:126–136. - PubMed
-
- Ashley, T., C. Westphal, A. Plug-de Maggio, and D.G. de Rooij. 2004. b. The mammalian mid-pachytene checkpoint: meiotic arrest in spermatocytes with a mutation in Atm alone or in combination with a Trp53 (p53) or Cdkn1a (p21/cip1) mutation. Cytogenet. Genome Res. 107:256–262. - PubMed
-
- Barchi, M., S. Mahadevaiah, M. Di Giacomo, F. Baudat, D.G. de Rooij, P.S. Burgoyne, M. Jasin, and S. Keeney. 2005. Surveillance of different recombination defects in mouse spermatocytes yields distinct responses despite elimination at an identical developmental stage. Mol. Cell. Biol. 25:7203–7215. - PMC - PubMed
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Miscellaneous

