The blue-light photoreceptor CRYPTOCHROME is expressed in a subset of circadian oscillator neurons in the Drosophila CNS
- PMID: 18663237
- PMCID: PMC2536721
- DOI: 10.1177/0748730408318588
The blue-light photoreceptor CRYPTOCHROME is expressed in a subset of circadian oscillator neurons in the Drosophila CNS
Abstract
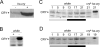
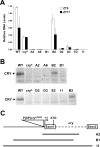
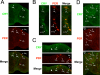
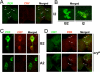
In the fruit fly Drosophila melanogaster, CRYPTOCHROME (CRY) functions as a photoreceptor to entrain circadian oscillators to light-dark cycles and as a transcription factor to maintain circadian oscillator function in certain peripheral tissues. Given the importance of CRY to circadian clock function, we expected this protein to be expressed in all oscillator cells, yet CRY cellular distribution and subcellular localization has not been firmly established. Here we investigate CRY spatial expression in the brain using a newly developed CRY antibody and a novel set of cry deletion mutants. We find that CRY is expressed in s-LNvs, l-LNvs, and a subset of LNds and DN1s, but not DN2s and DN3s. CRY is present in both the nucleus and the cytoplasm of these neurons, and its subcellular localization does not change over the circadian cycle. Although CRY is absent in DN2s and DN3s, cry promoter activity and/or cry mRNA accumulation can be detected in these neurons, suggesting that CRY levels are regulated posttranscriptionally. Oscillators in DN2s and DN3s entrain to environmental light-dark cycles, which implies that they are entrained indirectly by retinal photoreceptors, extraretinal photoreceptors, or other CRY-expressing cells.
Figures
Similar articles
-
Novel features of cryptochrome-mediated photoreception in the brain circadian clock of Drosophila.J Neurosci. 2004 Feb 11;24(6):1468-77. doi: 10.1523/JNEUROSCI.3661-03.2004. J Neurosci. 2004. PMID: 14960620 Free PMC article.
-
Spatial and circadian regulation of cry in Drosophila.J Biol Rhythms. 2008 Aug;23(4):283-95. doi: 10.1177/0748730408318566. J Biol Rhythms. 2008. PMID: 18663236 Free PMC article.
-
Circadian photoreception in Drosophila: functions of cryptochrome in peripheral and central clocks.J Biol Rhythms. 2001 Jun;16(3):205-15. doi: 10.1177/074873040101600303. J Biol Rhythms. 2001. PMID: 11407780
-
A fly's eye view of circadian entrainment.J Biol Rhythms. 2003 Jun;18(3):206-16. doi: 10.1177/0748730403018003003. J Biol Rhythms. 2003. PMID: 12828278 Review.
-
Structure and function of animal cryptochromes.Cold Spring Harb Symp Quant Biol. 2007;72:119-31. doi: 10.1101/sqb.2007.72.015. Cold Spring Harb Symp Quant Biol. 2007. PMID: 18419269 Review.
Cited by
-
Light/Clock Influences Membrane Potential Dynamics to Regulate Sleep States.Front Neurol. 2021 Mar 29;12:625369. doi: 10.3389/fneur.2021.625369. eCollection 2021. Front Neurol. 2021. PMID: 33854471 Free PMC article. Review.
-
Circadian rhythm of temperature preference and its neural control in Drosophila.Curr Biol. 2012 Oct 9;22(19):1851-7. doi: 10.1016/j.cub.2012.08.006. Epub 2012 Sep 13. Curr Biol. 2012. PMID: 22981774 Free PMC article.
-
Better Sleep at Night: How Light Influences Sleep in Drosophila.Front Physiol. 2020 Sep 4;11:997. doi: 10.3389/fphys.2020.00997. eCollection 2020. Front Physiol. 2020. PMID: 33013437 Free PMC article. Review.
-
PDFR and CRY signaling converge in a subset of clock neurons to modulate the amplitude and phase of circadian behavior in Drosophila.PLoS One. 2011 Apr 29;6(4):e18974. doi: 10.1371/journal.pone.0018974. PLoS One. 2011. PMID: 21559487 Free PMC article.
-
A Distinct Visual Pathway Mediates High-Intensity Light Adaptation of the Circadian Clock in Drosophila.J Neurosci. 2019 Feb 27;39(9):1621-1630. doi: 10.1523/JNEUROSCI.1497-18.2018. Epub 2019 Jan 3. J Neurosci. 2019. PMID: 30606757 Free PMC article.
References
-
- Busza A, Emery-Le M, Rosbash M, Emery P. Roles of the two Drosophila CRYPTOCHROME structural domains in circadian photoreception. Science. 2004;304:1503–1506. - PubMed
-
- Cashmore AR, Jarillo JA, Wu YJ, Liu D. Cryptochromes: blue light receptors for plants and animals. Science. 1999;284:760–765. - PubMed
-
- Ceriani MF, Darlington TK, Staknis D, Mas P, Petti AA, Weitz CJ, Kay SA. Light-dependent sequestration of TIMELESS by CRYPTOCHROME. Science. 1999;285:553–556. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

