Arabidopsis PUB22 and PUB23 are homologous U-Box E3 ubiquitin ligases that play combinatory roles in response to drought stress
- PMID: 18664614
- PMCID: PMC2518226
- DOI: 10.1105/tpc.108.060699
Arabidopsis PUB22 and PUB23 are homologous U-Box E3 ubiquitin ligases that play combinatory roles in response to drought stress
Abstract
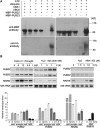
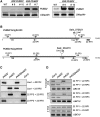
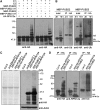
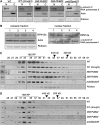
Ubiquitination is involved in diverse cellular processes in higher plants. In this report, we describe Arabidopsis thaliana PUB22 and PUB23, two homologous U-box-containing E3 ubiquitin (Ub) ligases. The PUB22 and PUB23 genes were rapidly and coordinately induced by abiotic stresses but not by abscisic acid. PUB22- and PUB23-overexpressing transgenic plants were hypersensitive to drought stress. By contrast, loss-of-function pub22 and pub23 mutant plants were significantly more drought-tolerant, and a pub22 pub23 double mutant displayed even greater drought tolerance. These results indicate that PUB22 and PUB23 function as negative regulators in the water stress response. Yeast two-hybrid, in vitro pull-down, and in vivo coimmunoprecipitation experiments revealed that PUB22 and PUB23 physically interacted with RPN12a, a subunit of the 19S regulatory particle (RP) in the 26S proteasome. Bacterially expressed RPN12a was effectively ubiquitinated in a PUB-dependent fashion. RPN12a was highly ubiquitinated in 35S:PUB22 plants, but not in pub22 pub23 double mutant plants, consistent with RPN12a being a substrate of PUB22 and PUB23 in vivo. In water-stressed wild-type and PUB-overexpressing plants, a significant amount of RPN12a was dissociated from the 19S RP and appeared to be associated with small-molecular-mass protein complexes in cytosolic fractions, where PUB22 and PUB23 are localized. Overall, our results suggest that PUB22 and PUB23 coordinately control a drought signaling pathway by ubiquitinating cytosolic RPN12a in Arabidopsis.
Figures




References
-
- Amador, V., Monte, E., García-Martínez, J.L., and Prat, S. (2001). Gibberellins signal nuclear import of PHOR1, a photoperiod-responsive protein with homology to Drosophila armadillo. Cell 106 343–354. - PubMed
-
- Andersen, P., Kragelund, B.B., Olsen, A.N., Larsen, F.H., Chua, N.-H., Poulsen, F.M., and Skriver, K. (2004). Structure and biochemical function of a prototypical Arabidopsis U-box domain. J. Biol. Chem. 279 40053–40061. - PubMed
-
- Azevedo, C., Santos-Rosa, M.J., and Shirasu, K. (2001). The U-box protein family in plants. Trends Plant Sci. 6 354–358. - PubMed
-
- Baumeister, W., Walz, J., Zuhl, F., and Seemuller, E. (1998). The proteasome: Paradigm of a self-compartmentalizing protease. Cell 92 367–380. - PubMed
-
- Boyer, J.S. (1982). Plant productivity and environment. Science 218 443–448. - PubMed
Publication types
MeSH terms
Substances
Associated data
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

