Global analysis of the medulloblastoma epigenome identifies disease-subgroup-specific inactivation of COL1A2
- PMID: 18664619
- PMCID: PMC2719012
- DOI: 10.1215/15228517-2008-048
Global analysis of the medulloblastoma epigenome identifies disease-subgroup-specific inactivation of COL1A2
Abstract
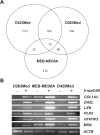
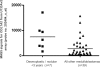
Candidate gene investigations have indicated a significant role for epigenetic events in the pathogenesis of medulloblastoma, the most common malignant brain tumor of childhood. To assess the medulloblastoma epigenome more comprehensively, we undertook a genomewide investigation to identify genes that display evidence of methylation-dependent regulation. Expression microarray analysis of medulloblastoma cell lines following treatment with a DNA methyltransferase inhibitor revealed deregulation of multiple transcripts (3%-6% of probes per cell line). Eighteen independent genes demonstrated >3-fold reactivation in all cell lines tested. Bisulfite sequence analysis revealed dense CpG island methylation associated with transcriptional silencing for 12 of these genes. Extension of this analysis to primary tumors and the normal cerebellum revealed three major classes of epigenetically regulated genes: (1) normally methylated genes (DAZL, ZNF157, ASN) whose methylation reflects somatic patterns observed in the cerebellum, (2) X-linked genes (MSN, POU3F4, HTR2C) that show disruption of their sex-specific methylation patterns in tumors, and (3) tumor-specific methylated genes (COL1A2, S100A10, S100A6, HTATIP2, CDH1, LXN) that display enhanced methylation levels in tumors compared with the cerebellum. Detailed analysis of COL1A2 supports a key role in medulloblastoma tumorigenesis; dense biallelic methylation associated with transcriptional silencing was observed in 46 of 60 cases (77%). Moreover, COL1A2 status distinguished infant medulloblastomas of the desmoplastic histopathological subtype, indicating that distinct molecular pathogenesis may underlie these tumors and their more favorable prognosis. These data reveal a more diverse and expansive medulloblastoma epi genome than previously understood and provide strong evidence that the methylation status of specific genes may contribute to the biological subclassification of medulloblastoma.
Figures



References
-
- Ellison DW, Clifford SC, Gajjar A, Gilbertson RJ. What’s new in neuro-oncology? Recent advances in medulloblastoma. Eur J Paediatr Neurol. 2003;7:53–66. - PubMed
-
- Ellison DW, Clifford SC, Giangaspero F. Medulloblastoma. In: McLendon RE, Rosenblum MK, Bigner DD, editors. Russell and Rubenstein’s Pathology of Tumours of the Nervous System. 7th ed. London: Hodder Arnold; 2006. pp. 247–264.
-
- Ellison DW, Onilude OE, Lindsey JC, et al. beta-Catenin status predicts a favorable outcome in childhood medulloblastoma: the United Kingdom Children’s Cancer Study Group Brain Tumour Committee. J Clin Oncol. 2005;23:7951–7957. - PubMed
-
- Thompson MC, Fuller C, Hogg TL, et al. Genomics identifies medulloblastoma subgroups that are enriched for specific genetic alterations. J Clin Oncol. 2006;24:1924–1931. - PubMed
-
- McManamy CS, Lamont JM, Taylor RE, et al. Morphophenotypic variation predicts clinical behavior in childhood non-desmoplastic medulloblastomas. J Neuropathol Exp Neurol. 2003;62:627–632. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials
Miscellaneous

