The N-terminal domain of the Drosophila retinoblastoma protein Rbf1 interacts with ORC and associates with chromatin in an E2F independent manner
- PMID: 18665226
- PMCID: PMC2475671
- DOI: 10.1371/journal.pone.0002831
The N-terminal domain of the Drosophila retinoblastoma protein Rbf1 interacts with ORC and associates with chromatin in an E2F independent manner
Abstract
Background: The retinoblastoma (Rb) tumor suppressor protein can function as a DNA replication inhibitor as well as a transcription factor. Regulation of DNA replication may occur through interaction of Rb with the origin recognition complex (ORC).
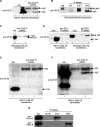
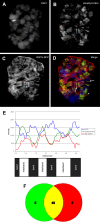
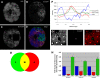
Principal findings: We characterized the interaction of Drosophila Rb, Rbf1, with ORC. Using expression of proteins in Drosophila S2 cells, we found that an N-terminal Rbf1 fragment (amino acids 1-345) is sufficient for Rbf1 association with ORC but does not bind to dE2F1. We also found that the C-terminal half of Rbf1 (amino acids 345-845) interacts with ORC. We observed that the amino-terminal domain of Rbf1 localizes to chromatin in vivo and associates with chromosomal regions implicated in replication initiation, including colocalization with Orc2 and acetylated histone H4.
Conclusions/significance: Our results suggest that Rbf1 can associate with ORC and chromatin through domains independent of the E2F binding site. We infer that Rbf1 may play a role in regulating replication directly through its association with ORC and/or chromatin factors other than E2F. Our data suggest an important role for retinoblastoma family proteins in cell proliferation and tumor suppression through interaction with the replication initiation machinery.
Conflict of interest statement
Figures




References
-
- Machida YJ, Hamlin JL, Dutta A. Right place, right time, and only once: Replication initiation in metazoans. Cell. 2005;123(1):13–24. - PubMed
-
- Diffley JF. Once and only once upon a time: Specifying and regulating origins of DNA replication in eukaryotic cells. Genes Dev. 1996;10(22):2819–2830. - PubMed
-
- Knudsen ES, Sexton CR, Mayhew CN. Role of the retinoblastoma tumor suppressor in the maintenance of genome integrity. Curr Mol Med. 2006;6(7):749–757. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

