Characterization of a modular enzyme of exo-1,5-alpha-L-arabinofuranosidase and arabinan binding module from Streptomyces avermitilis NBRC14893
- PMID: 18665359
- PMCID: PMC2518083
- DOI: 10.1007/s00253-008-1551-x
Characterization of a modular enzyme of exo-1,5-alpha-L-arabinofuranosidase and arabinan binding module from Streptomyces avermitilis NBRC14893
Abstract
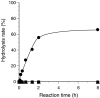
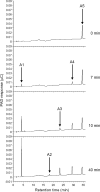
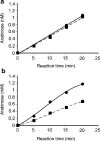
A gene encoding an alpha-L-arabinofuranosidase, designated SaAraf43A, was cloned from Streptomyces avermitilis. The deduced amino acid sequence implies a modular structure consisting of an N-terminal glycoside hydrolase family 43 module and a C-terminal family 42 carbohydrate-binding module (CBM42). The recombinant enzyme showed optimal activity at pH 6.0 and 45 degrees C and was stable over the pH range of 5.0-6.5 at 30 degrees C. The enzyme hydrolyzed p-nitrophenol (PNP)-alpha-L-arabinofuranoside but did not hydrolyze PNP-alpha-L-arabinopyranoside, PNP-beta-D-xylopyranoside, or PNP-beta-D-galactopyranoside. Debranched 1,5-arabinan was hydrolyzed by the enzyme but arabinoxylan, arabinogalactan, gum arabic, and arabinan were not. Among the synthetic regioisomers of arabinofuranobiosides, only methyl 5-O-alpha-L-arabinofuranosyl-alpha-L-arabinofuranoside was hydrolyzed by the enzyme, while methyl 2-O-alpha-L-arabinofuranosyl-alpha-L-arabinofuranoside and methyl 3-O-alpha-L-arabinofuranosyl-alpha-L-arabinofuranoside were not. These data suggested that the enzyme only cleaves alpha-1,5-linked arabinofuranosyl linkages. The analysis of the hydrolysis product of arabinofuranopentaose suggested that the enzyme releases arabinose in exo-acting manner. These results indicate that the enzyme is definitely an exo-1,5-alpha-L-arabinofuranosidase. The C-terminal CBM42 did not show any affinity for arabinogalactan and debranched arabinan, although it bound arabinan and arabinoxylan, suggesting that the CBM42 bound to branched arabinofuranosyl residues. Removal of the module decreased the activity of the enzyme with regard to debranched arabinan. The CBM42 plays a role in enhancing the debranched arabinan hydrolytic action of the catalytic module in spite of its preference for binding arabinofuranosyl side chains.
Figures




Similar articles
-
Two Novel α-l-Arabinofuranosidases from Bifidobacterium longum subsp. longum Belonging to Glycoside Hydrolase Family 43 Cooperatively Degrade Arabinan.Appl Environ Microbiol. 2019 Mar 6;85(6):e02582-18. doi: 10.1128/AEM.02582-18. Print 2019 Mar 15. Appl Environ Microbiol. 2019. PMID: 30635377 Free PMC article.
-
Purification, characterization and gene cloning of two alpha-L-arabinofuranosidases from streptomyces chartreusis GS901.Biochem J. 2000 Feb 15;346 Pt 1(Pt 1):9-15. Biochem J. 2000. PMID: 10657233 Free PMC article.
-
Substrate specificity of the alpha-L-arabinofuranosidase from Trichoderma reesei.Biosci Biotechnol Biochem. 1998 Nov;62(11):2205-10. doi: 10.1271/bbb.62.2205. Biosci Biotechnol Biochem. 1998. PMID: 9972241
-
Structure and function of carbohydrate-binding module families 13 and 42 of glycoside hydrolases, comprising a β-trefoil fold.Biosci Biotechnol Biochem. 2013;77(7):1363-71. doi: 10.1271/bbb.130183. Epub 2013 Jul 7. Biosci Biotechnol Biochem. 2013. PMID: 23832347 Review.
-
Exo- and Endo-1,5-α-L-Arabinanases and Prebiotic Arabino-Oligosaccharides Production.J Microbiol Biotechnol. 2025 Jan 13;35:e2412052. doi: 10.4014/jmb.2412.12052. J Microbiol Biotechnol. 2025. PMID: 39849927 Free PMC article. Review.
Cited by
-
Two Novel α-l-Arabinofuranosidases from Bifidobacterium longum subsp. longum Belonging to Glycoside Hydrolase Family 43 Cooperatively Degrade Arabinan.Appl Environ Microbiol. 2019 Mar 6;85(6):e02582-18. doi: 10.1128/AEM.02582-18. Print 2019 Mar 15. Appl Environ Microbiol. 2019. PMID: 30635377 Free PMC article.
-
Crystal structure and characterization of the glycoside hydrolase family 62 α-L-arabinofuranosidase from Streptomyces coelicolor.J Biol Chem. 2014 Mar 14;289(11):7962-72. doi: 10.1074/jbc.M113.540542. Epub 2014 Jan 30. J Biol Chem. 2014. PMID: 24482228 Free PMC article.
-
Two Distinct α-l-Arabinofuranosidases in Caldicellulosiruptor Species Drive Degradation of Arabinose-Based Polysaccharides.Appl Environ Microbiol. 2017 Jun 16;83(13):e00574-17. doi: 10.1128/AEM.00574-17. Print 2017 Jul 1. Appl Environ Microbiol. 2017. PMID: 28432102 Free PMC article.
-
A beta-l-Arabinopyranosidase from Streptomyces avermitilis is a novel member of glycoside hydrolase family 27.J Biol Chem. 2009 Sep 11;284(37):25097-106. doi: 10.1074/jbc.M109.022723. Epub 2009 Jul 16. J Biol Chem. 2009. PMID: 19608743 Free PMC article.
-
Overexpression, crystallization and preliminary X-ray crystallographic analysis of glucuronoxylan xylanohydrolase (Xyn30A) from Clostridium thermocellum.Acta Crystallogr Sect F Struct Biol Cryst Commun. 2013 Dec;69(Pt 12):1440-2. doi: 10.1107/S1744309113025050. Epub 2013 Nov 30. Acta Crystallogr Sect F Struct Biol Cryst Commun. 2013. PMID: 24316849 Free PMC article.
References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Miscellaneous

