Early indicators of exposure to biological threat agents using host gene profiles in peripheral blood mononuclear cells
- PMID: 18667072
- PMCID: PMC2542375
- DOI: 10.1186/1471-2334-8-104
Early indicators of exposure to biological threat agents using host gene profiles in peripheral blood mononuclear cells
Abstract
Background: Effective prophylaxis and treatment for infections caused by biological threat agents (BTA) rely upon early diagnosis and rapid initiation of therapy. Most methods for identifying pathogens in body fluids and tissues require that the pathogen proliferate to detectable and dangerous levels, thereby delaying diagnosis and treatment, especially during the prelatent stages when symptoms for most BTA are indistinguishable flu-like signs.
Methods: To detect exposures to the various pathogens more rapidly, especially during these early stages, we evaluated a suite of host responses to biological threat agents using global gene expression profiling on complementary DNA arrays.
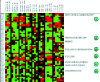
Results: We found that certain gene expression patterns were unique to each pathogen and that other gene changes occurred in response to multiple agents, perhaps relating to the eventual course of illness. Nonhuman primates were exposed to some pathogens and the in vitro and in vivo findings were compared. We found major gene expression changes at the earliest times tested post exposure to aerosolized B. anthracis spores and 30 min post exposure to a bacterial toxin.
Conclusion: Host gene expression patterns have the potential to serve as diagnostic markers or predict the course of impending illness and may lead to new stage-appropriate therapeutic strategies to ameliorate the devastating effects of exposure to biothreat agents.
Figures







Similar articles
-
Cluster analysis of host cytokine responses to biodefense pathogens in a whole blood ex vivo exposure model (WEEM).BMC Microbiol. 2012 May 20;12:79. doi: 10.1186/1471-2180-12-79. BMC Microbiol. 2012. PMID: 22607329 Free PMC article.
-
Modeling of SEB-induced host gene expression to correlate in vitro to in vivo responses.Biosens Bioelectron. 2004 Nov 1;20(4):719-27. doi: 10.1016/j.bios.2004.06.043. Biosens Bioelectron. 2004. PMID: 15522586
-
Activity of the Bacillus anthracis 20 kDa protective antigen component.BMC Infect Dis. 2008 Sep 22;8:124. doi: 10.1186/1471-2334-8-124. BMC Infect Dis. 2008. PMID: 18808698 Free PMC article.
-
Biological threat detection via host gene expression profiling.Clin Chem. 2003 Jul;49(7):1045-9. doi: 10.1373/49.7.1045. Clin Chem. 2003. PMID: 12816899 Review.
-
Pathology of inhalational anthrax animal models.Vet Pathol. 2010 Sep;47(5):819-30. doi: 10.1177/0300985810378112. Epub 2010 Jul 23. Vet Pathol. 2010. PMID: 20656900 Review.
Cited by
-
Transcriptome characterization of immune suppression from battlefield-like stress.Genes Immun. 2013 Jan;14(1):19-34. doi: 10.1038/gene.2012.49. Epub 2012 Oct 25. Genes Immun. 2013. PMID: 23096155 Free PMC article.
-
Protection of macrophages from intracellular pathogens by miR-182-5p mimic-a gene expression meta-analysis approach.FEBS J. 2018 Jan;285(2):244-260. doi: 10.1111/febs.14348. Epub 2017 Dec 26. FEBS J. 2018. PMID: 29197182 Free PMC article.
-
Host-Based Peripheral Blood Gene Expression Analysis for Diagnosis of Infectious Diseases.J Clin Microbiol. 2017 Feb;55(2):360-368. doi: 10.1128/JCM.01057-16. Epub 2016 Oct 19. J Clin Microbiol. 2017. PMID: 27795332 Free PMC article. Review.
-
Pathogen class-specific transcriptional responses derived from PBMCs accurately discriminate between fungal, bacterial, and viral infections.PLoS One. 2024 Dec 12;19(12):e0311007. doi: 10.1371/journal.pone.0311007. eCollection 2024. PLoS One. 2024. PMID: 39666613 Free PMC article.
-
Cluster analysis of host cytokine responses to biodefense pathogens in a whole blood ex vivo exposure model (WEEM).BMC Microbiol. 2012 May 20;12:79. doi: 10.1186/1471-2180-12-79. BMC Microbiol. 2012. PMID: 22607329 Free PMC article.
References
-
- Henchal EA, Teska JD, Ludwig GV, Shoemaker DR, Ezzell JW. Current laboratory methods for biological threat agent identification. Clin Lab Med. 2001;21:661–678. - PubMed
-
- Sun P, Celluzzi CM, Marovich M, Subramanian H, Eller M, Widjaja S, Palmer D, Porter K, Sun W, Burgess T. CD40 ligand enhances dengue viral infection of dendritic cells: a possible mechanism for T cell-mediated immunopathology. J Immunol. 2006;177:6497–6503. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

