The DNA damage sensors ataxia-telangiectasia mutated kinase and checkpoint kinase 2 are required for hepatitis C virus RNA replication
- PMID: 18667510
- PMCID: PMC2546985
- DOI: 10.1128/JVI.00351-08
The DNA damage sensors ataxia-telangiectasia mutated kinase and checkpoint kinase 2 are required for hepatitis C virus RNA replication
Abstract
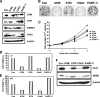
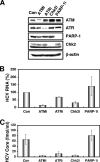
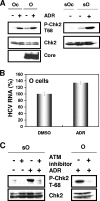
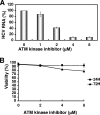
Cellular responses to DNA damage are crucial for maintaining genome integrity, virus infection, and preventing the development of cancer. Hepatitis C virus (HCV) infection and the expression of the HCV nonstructural protein NS3 and core protein have been proposed as factors involved in the induction of double-stranded DNA breaks and enhancement of the mutation frequency of cellular genes. Since DNA damage sensors, such as the ataxia-telangiectasia mutated kinase (ATM), ATM- and Rad3-related kinase (ATR), poly(ADP-ribose) polymerase 1 (PARP-1), and checkpoint kinase 2 (Chk2), play central roles in the response to genotoxic stress, we hypothesized that these sensors might affect HCV replication. To test this hypothesis, we examined the level of HCV RNA in HuH-7-derived cells stably expressing short hairpin RNA targeted to ATM, ATR, PARP-1, or Chk2. Consequently, we found that replication of both genome-length HCV RNA (HCV-O, genotype 1b) and the subgenomic replicon RNA were notably suppressed in ATM- or Chk2-knockdown cells. In addition, the RNA replication of HCV-JFH1 (genotype 2a) and the release of core protein into the culture supernatants were suppressed in these knockdown cells after inoculation of the cell culture-generated HCV. Consistent with these observations, ATM kinase inhibitor could suppress the HCV RNA replication. Furthermore, we observed that HCV NS3-NS4A interacted with ATM and that HCV NS5B interacted with both ATM and Chk2. Taken together, these results suggest that the ATM signaling pathway is critical for HCV RNA replication and may represent a novel target for the clinical treatment of patients with chronic hepatitis C.
Figures

References
-
- Bridge, A. J., S. Pebernard, A. Ducraux, A.-L. Nicoulaz, and R. Iggo. 2003. Induction of an interferon response by RNAi vectors in mammalian cells. Nat. Genet. 34263-264. - PubMed
-
- Brummelkamp, T. R., R. Bernard, and R. Agami. 2002. A system for stable expression of short interfering RNAs in mammalian cells. Science 296550-553. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials
Miscellaneous

