Nse1 RING-like domain supports functions of the Smc5-Smc6 holocomplex in genome stability
- PMID: 18667531
- PMCID: PMC2555936
- DOI: 10.1091/mbc.e08-02-0226
Nse1 RING-like domain supports functions of the Smc5-Smc6 holocomplex in genome stability
Abstract
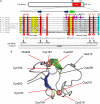
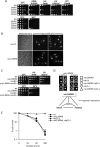
The Smc5-Smc6 holocomplex plays essential but largely enigmatic roles in chromosome segregation, and facilitates DNA repair. The Smc5-Smc6 complex contains six conserved non-SMC subunits. One of these, Nse1, contains a RING-like motif that often confers ubiquitin E3 ligase activity. We have functionally characterized the Nse1 RING-like motif, to determine its contribution to the chromosome segregation and DNA repair roles of Smc5-Smc6. Strikingly, whereas a full deletion of nse1 is lethal, the Nse1 RING-like motif is not essential for cellular viability. However, Nse1 RING mutant cells are hypersensitive to a broad spectrum of genotoxic stresses, indicating that the Nse1 RING motif promotes DNA repair functions of Smc5-Smc6. We tested the ability of both human and yeast Nse1 to mediate ubiquitin E3 ligase activity in vitro and found no detectable activity associated with full-length Nse1 or the isolated RING domains. Interestingly, however, the Nse1 RING-like domain is required for normal Nse1-Nse3-Nse4 trimer formation in vitro and for damage-induced recruitment of Nse4 and Smc5 to subnuclear foci in vivo. Thus, we propose that the Nse1 RING-like motif is a protein-protein interaction domain required for Smc5-Smc6 holocomplex integrity and recruitment to, or retention at, DNA lesions.
Figures



References
-
- Aravind L., Iyer L. M., Koonin E. V. Scores of RINGS but no PHDs in ubiquitin signaling. Cell Cycle. 2003;2:123–126. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

