Conformational flexibility facilitates self-assembly of complex DNA nanostructures
- PMID: 18667705
- PMCID: PMC2504817
- DOI: 10.1073/pnas.0803841105
Conformational flexibility facilitates self-assembly of complex DNA nanostructures
Abstract
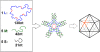
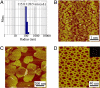
Molecular self-assembly is a promising approach to the preparation of nanostructures. DNA, in particular, shows great potential to be a superb molecular system. Synthetic DNA molecules have been programmed to assemble into a wide range of nanostructures. It is generally believed that rigidities of DNA nanomotifs (tiles) are essential for programmable self-assembly of well defined nanostructures. Recently, we have shown that adequate conformational flexibility could be exploited for assembling 3D objects, including tetrahedra, dodecahedra, and buckyballs, out of DNA three-point star motifs. In the current study, we have integrated tensegrity principle into this concept to assemble well defined, complex nanostructures in both 2D and 3D. A symmetric five-point-star motif (tile) has been designed to assemble into icosahedra or large nanocages depending on the concentration and flexibility of the DNA tiles. In both cases, the DNA tiles exhibit significant flexibilities and undergo substantial conformational changes, either symmetrically bending out of the plane or asymmetrically bending in the plane. In contrast to the complicated natures of the assembled structures, the approach presented here is simple and only requires three different component DNA strands. These results demonstrate that conformational flexibility could be explored to generate complex DNA nanostructures. The basic concept might be further extended to other biomacromolecular systems, such as RNA and proteins.
Conflict of interest statement
The authors declare no conflict of interest.
Figures
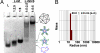


References
-
- Hamley IW. Nanotechnology with soft materials. Angew Chem Int Ed. 2003;42:1692–1712. - PubMed
-
- Reinhoudt DN, Crego-Calama M. Synthesis beyond the molecule. Science. 2002;295:2403–2407. - PubMed
-
- Seeman NC. DNA in a material world. Nature. 2003;421:427–431. - PubMed
-
- Winfree E, Liu FR, Wenzler LA, Seeman NC. Design and self-assembly of two-dimensional DNA crystals. Nature. 1998;394:539–544. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

