Global organization of replication time zones of the mouse genome
- PMID: 18669478
- PMCID: PMC2556267
- DOI: 10.1101/gr.079566.108
Global organization of replication time zones of the mouse genome
Abstract
The division of genomes into distinct replication time zones has long been established. However, an in-depth understanding of their organization and their relationship to transcription is incomplete. Taking advantage of a novel synchronization method ("baby machine") and of genomic DNA microarrays, we have, for the first time, mapped replication times of the entire mouse genome at a high temporal resolution. Our data revealed that although most of the genome has a distinct time of replication either early, middle, or late S phase, a significant portion of the genome is replicated asynchronously. Analysis of the replication map revealed the genomic scale organization of the replication time zones. We found that the genomic regions between early and late replication time zones often consist of extremely large replicons. Analysis of the relationship between replication and transcription revealed that early replication is frequently correlated with the transcription potential of a gene and not necessarily with its actual transcriptional activity. These findings, along with the strong conservation found between replication timing in human and mouse genomes, emphasize the importance of replication timing in transcription regulation.
Figures
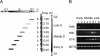


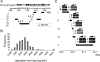

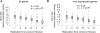
References
-
- Azuara V., Brown K.E., Williams R.R., Webb N., Dillon N., Festenstein R., Buckle V., Merkenschlager M., Fisher A.G., Brown K.E., Williams R.R., Webb N., Dillon N., Festenstein R., Buckle V., Merkenschlager M., Fisher A.G., Williams R.R., Webb N., Dillon N., Festenstein R., Buckle V., Merkenschlager M., Fisher A.G., Webb N., Dillon N., Festenstein R., Buckle V., Merkenschlager M., Fisher A.G., Dillon N., Festenstein R., Buckle V., Merkenschlager M., Fisher A.G., Festenstein R., Buckle V., Merkenschlager M., Fisher A.G., Buckle V., Merkenschlager M., Fisher A.G., Merkenschlager M., Fisher A.G., Fisher A.G. Heritable gene silencing in lymphocytes delays chromatid resolution without affecting the timing of DNA replication. Nat. Cell Biol. 2003;5:668–674. - PubMed
-
- Azuara V., Perry P., Sauer S., Spivakov M., Jorgensen H.F., John R.M., Gouti M., Casanova M., Warnes G., Merkenschlager M., Perry P., Sauer S., Spivakov M., Jorgensen H.F., John R.M., Gouti M., Casanova M., Warnes G., Merkenschlager M., Sauer S., Spivakov M., Jorgensen H.F., John R.M., Gouti M., Casanova M., Warnes G., Merkenschlager M., Spivakov M., Jorgensen H.F., John R.M., Gouti M., Casanova M., Warnes G., Merkenschlager M., Jorgensen H.F., John R.M., Gouti M., Casanova M., Warnes G., Merkenschlager M., John R.M., Gouti M., Casanova M., Warnes G., Merkenschlager M., Gouti M., Casanova M., Warnes G., Merkenschlager M., Casanova M., Warnes G., Merkenschlager M., Warnes G., Merkenschlager M., Merkenschlager M., et al. Chromatin signatures of pluripotent cell lines. Nat. Cell Biol. 2006;8:532–538. - PubMed
-
- Berezney R., Dubey D.D., Huberman J.A., Dubey D.D., Huberman J.A., Huberman J.A. Heterogeneity of eukaryotic replicons, replicon clusters, and replication foci. Chromosoma. 2000;108:471–484. - PubMed
-
- Bierne H., Michel B., Michel B. When replication forks stop. Mol. Microbiol. 1994;13:17–23. - PubMed
-
- Braunstein J.D., Schulze D., DelGiudice T., Furst A., Schildkraut C.L., Schulze D., DelGiudice T., Furst A., Schildkraut C.L., DelGiudice T., Furst A., Schildkraut C.L., Furst A., Schildkraut C.L., Schildkraut C.L. The temporal order of replication of murine immunoglobulin heavy chain constant region sequences corresponds to their linear order in the genome. Nucleic Acids Res. 1982;10:6887–6902. - PMC - PubMed
Publication types
MeSH terms
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
