Submergence-responsive MicroRNAs are potentially involved in the regulation of morphological and metabolic adaptations in maize root cells
- PMID: 18669574
- PMCID: PMC2701776
- DOI: 10.1093/aob/mcn129
Submergence-responsive MicroRNAs are potentially involved in the regulation of morphological and metabolic adaptations in maize root cells
Abstract
Background and aims: Anaerobic or low oxygen conditions occur when maize plants are submerged or subjected to flooding of the soil. Maize survival under low oxygen conditions is largely dependent on metabolic, physiological and morphological adaptation strategies; the regulation mechanisms of which remain unknown. MicroRNAs (miRNAs) play critical roles in the response to adverse biotic or abiotic stresses at the post-transcriptional level. The aim of this study was to understand submergence-responsive miRNAs and their potential roles in submerged maize roots.
Methods: A custom muParaflo microfluidic array containing plant miRNA (miRBase: http://microrna.sanger.ac.uk) probes was used to explore differentially expressed miRNAs. Small RNAs from treated roots were hybridized with the microarray. The targets and their cis-acting elements of small RNA were predicted and analysed by RT-PCR.
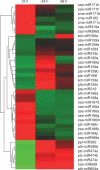
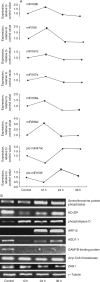
Key results: Microarray data revealed that the expression levels of 39 miRNAs from nine maize and some other plant miRNA families were significantly altered (P < 0.01). Four expression profiles were identified across different submergence time-points. The zma-miRNA166, zma-miRNA167, zma-miRNA171 and osa-miRNA396-like were induced in the early phase, and their target genes were predicted to encode important transcription factors, including; HD-ZIP, auxin response factor, SCL and the WRKY domain protein. zma-miR159, ath-miR395-like, ptc-miR474-like and osa-miR528-like were reduced at the early submergence phase and induced after 24 h of submergence. The predicted targets for these miRNAs were involved in carbohydrate and energy metabolism, including starch synthase, invertase, malic enzyme and ATPase. In addition, many of the predicted targets were involved in the elimination of reactive oxygen species and acetaldehyde. Overall, most of the targets of induced miRNAs contained the cis-acting element, which is essential for the anaerobic response or hormone induction.
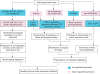
Conclusions: Submergence-responsive miRNAs are involved in the regulation of metabolic, physiological and morphological adaptations of maize roots at the post-transcriptional level.
Figures

Comment in
-
Transcriptional and post-transcriptional regulation of gene expression in submerged root cells of maize.Plant Signal Behav. 2009 Feb;4(2):132-5. doi: 10.4161/psb.4.2.7629. Plant Signal Behav. 2009. PMID: 19649190 Free PMC article.
Similar articles
-
Differential expression of miRNAs in response to salt stress in maize roots.Ann Bot. 2009 Jan;103(1):29-38. doi: 10.1093/aob/mcn205. Epub 2008 Oct 24. Ann Bot. 2009. PMID: 18952624 Free PMC article.
-
Genome-wide identification and analysis of microRNA responding to long-term waterlogging in crown roots of maize seedlings.Physiol Plant. 2013 Feb;147(2):181-93. doi: 10.1111/j.1399-3054.2012.01653.x. Epub 2012 Jun 27. Physiol Plant. 2013. PMID: 22607471
-
Genome-wide identification of microRNAs in response to low nitrate availability in maize leaves and roots.PLoS One. 2011;6(11):e28009. doi: 10.1371/journal.pone.0028009. Epub 2011 Nov 23. PLoS One. 2011. PMID: 22132192 Free PMC article.
-
Transcriptional regulatory networks in response to drought stress and rewatering in maize (Zea mays L.).Mol Genet Genomics. 2021 Nov;296(6):1203-1219. doi: 10.1007/s00438-021-01820-y. Epub 2021 Oct 3. Mol Genet Genomics. 2021. PMID: 34601650 Review.
-
MicroRNAs and their diverse functions in plants.Plant Mol Biol. 2012 Sep;80(1):17-36. doi: 10.1007/s11103-011-9817-6. Epub 2011 Aug 27. Plant Mol Biol. 2012. PMID: 21874378 Review.
Cited by
-
Consequences of waterlogging in cotton and opportunities for mitigation of yield losses.AoB Plants. 2015 Jul 20;7:plv080. doi: 10.1093/aobpla/plv080. AoB Plants. 2015. PMID: 26194168 Free PMC article.
-
Dual transcriptome analysis reveals insights into the response to Rice black-streaked dwarf virus in maize.J Exp Bot. 2016 Aug;67(15):4593-609. doi: 10.1093/jxb/erw244. Epub 2016 Jul 18. J Exp Bot. 2016. PMID: 27493226 Free PMC article.
-
Differential Expression of MicroRNAs During Root Formation in Taxus Chinensis Var. mairei Cultivars.Open Life Sci. 2019 Apr 6;14:97-109. doi: 10.1515/biol-2019-0011. eCollection 2019 Jan. Open Life Sci. 2019. PMID: 33817141 Free PMC article.
-
High Daytime Temperature Responsive MicroRNA Profiles in Developing Grains of Rice Varieties with Contrasting Chalkiness.Int J Mol Sci. 2023 Jul 19;24(14):11631. doi: 10.3390/ijms241411631. Int J Mol Sci. 2023. PMID: 37511395 Free PMC article.
-
Stress-responsive miRNAome of Glycine max (L.) Merrill: molecular insights and way forward.Planta. 2019 May;249(5):1267-1284. doi: 10.1007/s00425-019-03114-5. Epub 2019 Feb 23. Planta. 2019. PMID: 30798358 Review.
References
-
- Achard P, Herr A, Baulcombe DC, Harberd NP. Modulation of floral development by a gibberellin-regulated microRNA. Development. 2004;131:3357–3365. - PubMed
-
- Agarwal S, Grover A. Molecular biology, biotechnology and genomics of flooding-associated low O2 stress response in plant. Critical Reviews in Plant Sciences. 2006;25:1–21.
-
- Armstrong W. Radial oxygen losses from intact rice roots as affected by distance from the apex, respiration and waterlogging. Physiologia Plantarum. 1971;25:192–197.
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials
Miscellaneous

